Ribosomal RNA guanine-(N2)-methyltransferases and their targets (original) (raw)
Abstract
Five nearly universal methylated guanine-(N2) residues are present in bacterial rRNA in the ribosome. To date four out of five ribosomal RNA guanine-(N2)-methyltransferases are described. RsmC(YjjT) methylates G1207 of the 16S rRNA. RlmG(YgjO) and RlmL(YcbY) are responsible for the 23S rRNA m2G1835 and m2G2445 formation, correspondingly. RsmD(YhhF) is necessary for methylation of G966 residue of 16S rRNA. Structure of Escherichia coli RsmD(YhhF) methyltransferase and the structure of the Methanococcus jannaschii RsmC ortholog were determined. All ribosomal guanine-(N2)-methyltransferases have similar AdoMet-binding sites. In relation to the ribosomal substrate recognition, two enzymes that recognize assembled subunits are relatively small single domain proteins and two enzymes that recognize naked rRNA are larger proteins containing separate methyltransferase- and RNA-binding domains. The model for recognition of specific target nucleotide is proposed. The hypothetical role of the m2G residues in rRNA is discussed.
INTRODUCTION
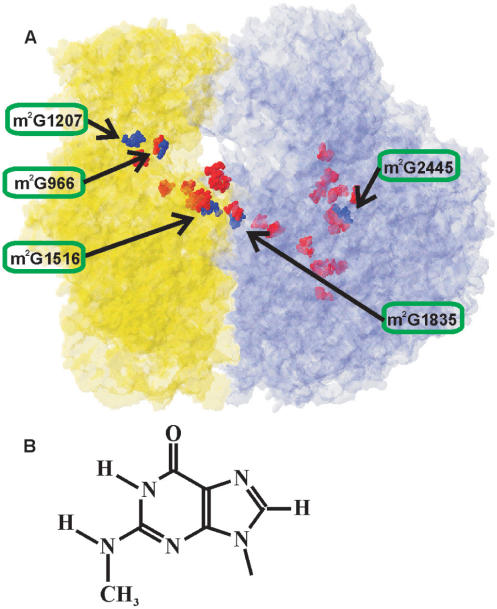
RNA composes a structural and functional core of the ribosome, the protein factory of the cell. Ribosomal RNA of Escherichia coli is represented with 120 nt-long 5S rRNA, and two large molecules: 16S rRNA of 1542 nt and 23S rRNA of 2904 nt. Among 4500 rRNA nucleotide residues, 36 are modified (1). In the tertiary structure of the ribosome, modified nucleotides are concentrated in a continuous cluster (Figure 1A), encompassing functional centers (2). The decoding center of the small subunit, the subunit interface and the peptidyltransferase center of large subunit are particularly enriched in the modified bases. In some cases, the role of ribosomal RNA modifications was established. These are modifications protecting some bacteria from antibiotics [see (3) for review]. They were found in the limited sets of organisms, such as antibiotic producers (3) and unfortunately some clinically important pathogens (4), among those spread of resistance was being caused by wide-scale antibiotic usage. Despite almost entire lack of information regarding the function of the remaining rRNA-modified nucleotides, their clustering in the functional centers suggests their importance in translation process. Here, we will discuss N2-methyl guanosine (m2G) modifications (Figure 1B), which were a point of our recent investigation.
Figure 1.
(A) Modified nucleotides distribution in the structure of the E. coli ribosome (27). Small subunit is shown in yellow; large subunit is shown in blue. N2-methylguanosine residues are shown as dark blue Van-der-Vaals spheres. The residue number of m2G nucleotides in 16S or 23S rRNA is depicted and the location of particular m2G residue is indicated by arrow. Other modified nucleotides are shown as red Van-der-Vaals spheres. (B) The structure of N2-methylguanine.
Among the modified nucleotides of bacteria, exemplified by E. coli, the most frequently found type is pseudouridines (1). There are 11 pseudouridines in the rRNA of E. coli, and all genes responsible for uridine to pseudouridine isomerization have been described (5). Here, we discuss the second most abundant modified nucleotide type of rRNA modification, namely, m2G. There are three m2G residues in the 16S rRNA and two in the 23S rRNA (1), whose positions are marked in blue in Figure 1. In 1999, the yjjT open reading frame subsequently being renamed to rsmC, was identified as encoding methyltransferase specific for the methylation of G1207 of the 16S rRNA (6). Several open reading frames were suggested to encode guanine-(N2)-methyltransferases on the basis of sequence analysis (7,8). However, only recently we were able to verify three genes responsible for m2G formation in rRNA experimentally. YhhF open reading frame was shown to encode RsmD methyltransferase, involved in the modification of G966 of the 16S rRNA (9). YgjO (10) and ycbY (11) open reading frames appeared to encode RlmG and RlmL enzymes modifying G1835 and G2445 of the 23S rRNA. Now, since four out of five ribosomal guanine-(N2)-methyltransferase genes were determined we can compare this distinctive class of enzymes, in an attempt to find their common features and differences.
STRUCTURES OF THE RIBOSOMAL GUANINE-(N2)-METHYLTRANSFERASES: IMPLICATIONS FOR SUBSTRATE SPECIFICITY
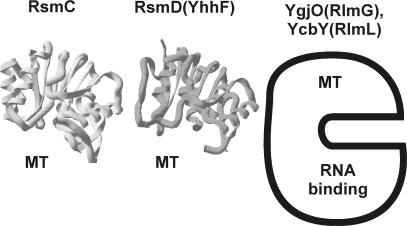
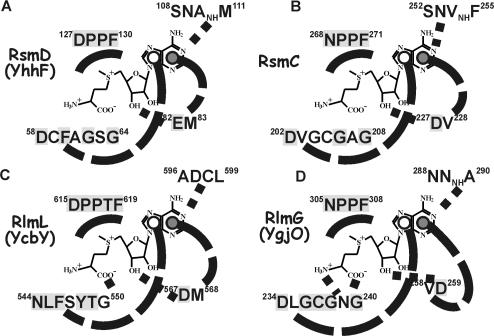
Of the four identified E. coli guanine-(N2)-methyltransferases, only RsmD(YhhF) tertiary structure has so far been determined by X-ray structure analysis (9). It consists of the single Rossman fold methyltransferase domain (Figure 2). The protein was crystallized without AdoMet, however the cofactor position could easily be reconstructed by superimposition of the known Rossman fold RNA methyltransferases structure with AdoMet bound. Binding pocket of the cofactor is formed by residues conserved among homologs of RsmD(YhhF) protein (Table S1) (marked gray on the Figure 3A). The core of AdoMet, surrounding the junction between methionine and adenosine should be sandwiched between 127DPPF130 and 58DxFxGxG64 motifs. Carboxyl and amino groups of methionine moiety of AdoMet are likely to form hydrogen bonds with aspartate and glycine of DxFxGxG, as in DpnM DNA methyltransferase. E82 is positioned in a way to interact with AdoMet ribose hydroxyl(s). Adenine base must fit between M83 and F60, while its Watson–Crick edge is likely to interact with 108SNAM111 protein fragment.
Figure 2.
The structure of ribosomal guanine-(N2)-methyltransferases. The structure of Mj0882 protein (12), which is likely to represent an ortholog of E. coli RsmC is marked as ‘RsmC’. The structure of E. coli RsmD(YhhF) protein is marked accordingly. Both protein structures are also marked ‘MT’ to indicate that they consist of single methyltransferase domain. For RlmG(YgjO) and RlmL(YcbY) the scheme is shown, where the protein is divided into N-terminal RNA-binding domain and C-terminal methyltransferase (MT) domain, based on conserved domain analysis (38).
Figure 3.
The scheme of AdoMet-binding site of the ribosomal guanine-(N2)-methyltransferases. (A) RsmD(YhhF) active site, (B) RsmC(YjjT) active site, (C) RlmL(YcbY) active site and (D) RlmG(YgjO) active site. AdoMet structure is shown in the center of each scheme. It is surrounded by protein sequences, proximal to the putative AdoMet-binding site. Conserved amino acids are shaded gray. Probable hydrogen bonds are indicated by dotted lines. Hypothetical stacking interactions of the adenine base are shown by dashed lines. Open circles indicate the location of amino acid side chain below the adenine plane; closed circles indicate that amino acid is stacked above the adenine plane.
Escherichia coli RsmC tertiary structure has not been solved yet, however Mj0882 protein of Methanococcus jannaschii (Figure 2), whose structure has been determined (12) is likely to represent RsmC ortholog (8). Reconstruction of E. coli RsmC AdoMet-binding site using sequence similarity with Mj0882 reveals overall architecture highly similar to that of RsmD(YhhF) (Figure 3B). Similarly, AdoMet core is located between 268NPPF271 and 202DxGxGxG208 conserved motifs. Conserved D227 is likely to form hydrogen bond with ribose hydroxyls of AdoMet, while adenine base should be stacked between V203 and V228.
Tertiary structure of RlmG(YgjO) and RlmL(YcbY) is unknown. Sequence comparisons lead to the conclusion that their AdoMet-binding site is constructed very similar to those of RsmC and RsmD(yhhF) (Figure 3C and D). Motifs 305NPPF308 and similar 615DPPTF619 with threonine single amino acid insertion are present in RlmG(YgjO) and RlmL(YcbY). Methionine part of AdoMet should be fitted between those proline-rich motifs and sequences 234DxGxGxG240 (YgjO) and 544NxFxYxG550 (YcbY). Aspartates 259(YgjO) and 567 (YcbY) are close to the putative position of AdoMet ribose hydroxyls. Adenine base is likely to be stacked between V258 and L235 (YgjO) and M568 and F546 (YcbY). Overall architecture of four E. coli rRNA guanine-(N2)-methyltransferases active site looks very similar. Despite this fact each of them should specifically recognize a single guanosine residue among ∼1000 guanosine residues of the ribosome.
Four E. coli rRNA guanine-(N2)-methyltransferases could be divided into two categories based on the domain structure and substrate specificity (Figure 2). RsmC(YjjT) and RsmD(YhhF) are relatively small proteins of 343 and 198 amino acid long. Both RsmC (6) and RsmD (9,13) could recognize assembled small ribosomal subunit as a substrate and perhaps their in vivo targets are late assembly intermediates resembling the completed 30S particle (13). RlmG and RlmL are bigger proteins 378 and 702 amino acid long, recognizing protein-free ribosomal RNA in vitro (10,11) and most probably, unfolded early assembly intermediates in vivo. Both RlmG and RlmL (Figure 2) have N-terminal RNA-binding domain in addition to conserved methyltransferase domain described above. Why some of guanine-(N2)-methyltransferases recognize naked rRNA while others could use assembled subunits? On the tertiary structure of the ribosome, nucleotides that are modified at early assembly stages are either buried in the core of the tertiary structure and became inaccessible after assembly (Figure 1; nucleotide m2G2445) or exposed on the surface making it difficult to distinguish from other surface-exposed guanines (Figure 1; nucleotide m2G1835). To provide the specificity for modification of particular G residue such enzymes should recognize additional RNA determinants. It becomes necessary to have additional RNA-binding domain and operate with partially unfolded rRNA. In contrast, both guanines G966 and G1207 are accessible for the respective enzymes and can be modified at amino group after subunit assembly despite their location in the deep decoding cleft of the small subunit (Figure 1; nucleotides m2G966 and m2G1207). Decoding cleft, according to the SwissPDBviewer-assisted (14) manual fitting of the RsmD(YhhF) structure (9) to the X-ray structure of Thermus thermophilus small subunit (15) (Figure 4) is sufficient to bind small methyltransferases (9). Thus, the problem of substrate recognition is solved by adjustment of enzyme outer surface to the shape of the cleft in the substrate that allows in achieving such active site positioning that only target G residue can be recognized. The ability of RsmC and RsmD enzymes to use natural cavity in the substrate to ensure sufficient specificity force them to be relatively small. Indeed these enzymes practically consist of only methyltransferase domain (Figure 2).
Figure 4.
Fitting of RsmD(YhhF) structure (dark gray) to the P-site of 3D structure of the small ribosomal subunit (light gray). T. thermophilus 30S subunit in open conformation (15), was used for the fitting. The head of the small subunit was moved upward by 4 Å to avoid steric clashes. This movement is highly probable in vivo given high conformational mobility of the small subunit head.
FUNCTION OF M2G RESIDUES IN THE RIBOSOMAL RNA
Modification of m2G residues is not of vital importance for the cell. However, lack of such modifications of the ribosomal RNA influence the cell growth at different conditions (9–11). The exact structural and functional role of the m2G residues can be hypothesized on the basis of their location in the ribosomal structure. The m2G residues in rRNA are located in the three clusters of modified nucleotides, coinciding with decoding and peptidyltransferase centers and subunit interface. Methylation of the amino group 2 of guanosine moiety (Figure 1B) could have two general consequences. First, addition of the hydrophobic group allows the formation of the hydrophobic contact with protein or RNA. This property might be important for m2G966, contacting tRNA anticodon. Second, addition of the methyl group eliminates one hydrogen bond donor. Amino group 2 is involved in the Watson–Crick interactions and has two hydrogen atoms. Monomethylation should not prevent complementary interaction of m2G with the cytosine of the opposite RNA strand, however base pair formation places methyl group unambiguously to the RNA minor groove. There it should prevent any hydrogen bond interactions, such as certain base triples. This might be the structural role of the G2445 methylation.
m2G 966 OF THE 16S rRNA
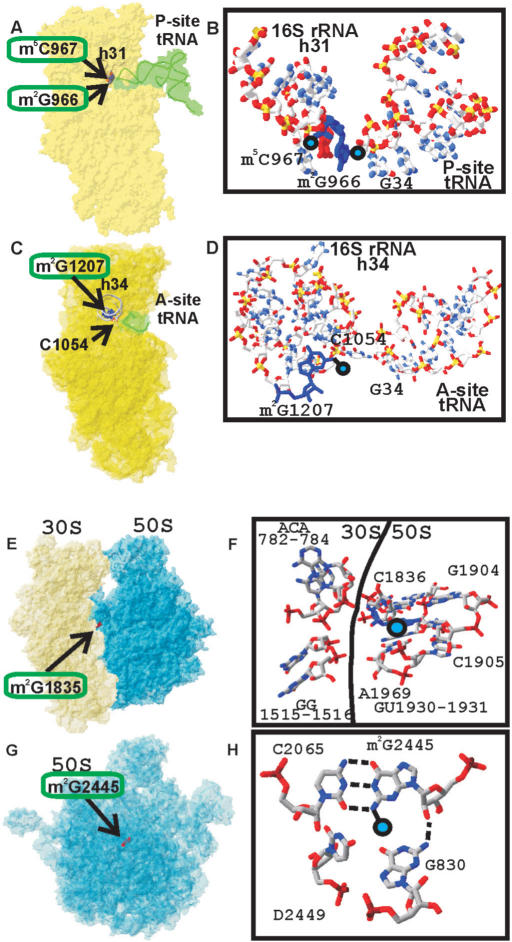
Nucleotide m2G966 is located in the loop of the helix 31 of the 16S rRNA next to the m5C967 (Figure 5A,B). Both modified nucleotides form a part of the binding surface for the tRNA anticodon in the P-site as was shown by footprinting in 1980s (16) and recently confirmed by X-ray structure analysis (17). Direct contact with the tip of tRNA anticodon might suggests that nucleotides 966–967 modifications to play important role, however several lines of evidence argues that this is not the case. Nucleotide 966 is not conserved among different kingdoms of life (18), however identity of this base is preserved within the kingdoms. In bacteria and chloroplasts nucleotide 966 is predominantly guanosine, while in archaea and eukarya it is uridine (18). Mitochondrial small subunit rRNA has adenosine at the position, equivalent to G966 (18). In agreement with lack of conservation in evolution, mutations of the nucleotide 966 do not have any severe phenotype (19). Curiously, in archaea and eukarya despite changing identity of the base 966 the modified character of this nucleotide is preserved. In archaea position 966 is occupied by acp3U (20), while in eukarya m1acp3ψ is located at this place (21).
Figure 5.
Location of the m2G966, m2G1207 residues of the 16S rRNA and m2G1835, m2G2445 residues of the 23S rRNA in the ribosomal structure and their interactions. (A) Position of the 16S rRNA residue m2G966 relative to the P-site-bound tRNA (17). The 30S subunit is shown in yellow. P-site-bound tRNA is shown in green. Helix 31 is indicated as a gray tubing. Modified nucleotides m2G966 and m2G967 are shown as a blue and red Van-der-Vaals spheres accordingly and labeled. (B) Closer view of the position of the 16S rRNA residue m2G966 relative to the P-site-bound tRNA. 16S rRNA helix 31 is shown on the left, P-site-bound tRNA anticodon is shown on the right. (C) Position of the 16S rRNA residue m2G1207 relative to the A-site-bound tRNA anticodon (23). The 30S subunit is shown in yellow. A-site-bound tRNA anticodon is shown in green. Helix 34 is indicated as a gray tubing. Modified nucleotide m2G1207 is shown as red Van-der-Vaals spheres and labeled. Nucleotide C1054 making a direct contact with A-site tRNA anticodon is shown as wireframe and labeled. (D) Closer view of the position of the 16S rRNA residue m2G1207 relative to the A-site-bound tRNA. 16S rRNA helix 34 is shown on the left, A-site-bound tRNA anticodon is shown on the right. (E) Position of the 23S rRNA residue m2G1835 in the E. coli ribosome (27). Position of the m2G1835 residue relative to the ribosomal subunits. The 30S subunit is shown in yellow; the 50S subunit is shown in blue. Modified nucleotide m2G1835 is shown as red Van-der-Vaals spheres and labeled. (F) Details of structural environment of the m2G1835 in the ribosome. Position of the methyl group is marked by blue circle. Surrounding nucleotides are shown as wireframe and labeled. 16S rRNA nucleotides are on the left side, 23S rRNA nucleotides are on the right side. Border between the subunits is indicated by line. (G) Position of the 23S rRNA residue m2G2445 in the E. coli 50S subunit (27). Position of the m2G2445 residue relative to the large ribosomal subunit, shown in blue and labeled. The orientation of the 50S subunit is as viewed from the 30S subunit. (H) Details of structural environment of the m2G2445 in the 50S subunit. Position of the methyl group is marked by blue circle. Surrounding nucleotides are shown as wireframe and labeled.
Deletion of yhhF gene in E. coli has almost no phenotype (9). Highly sensitive growth competition assay revealed very moderate growth disadvantage of the strain lacking G966 modification in comparison with the parental wild-type strain. Methylation of G966 and modification of this base in other kingdoms of life could prevent formation of some unproductive conformation of the small subunit rRNA. If the functional role of modification is not to cause the formation of some particular interaction, but to prevent formation of a particular interaction it explains why the identity of the base 966 is not so important. It seems that in the absence of methylation, rRNA could oscillate between functional and non-functional conformations, while methylation guarantees the preference for the functional one. If fully functional conformation is not dependent, but is only favoured by the G966 modification it explains why methylation is not so important. There is also a possibility that m2G966 is involved in binding of tRNA to the P-site. Binding of tRNA anticodon to the P-site of the small subunit is strong enough to make a single interaction with the methyl group dispensable. It is also possible that the function of m2G966 and m5C967 is redundant. Thus, a double knockout of rsmD and rsmB genes could have more severe phenotype.
m2G 1207 OF THE 16S rRNA
Nucleotide m2G1207 is located in the helix 34 of the 16S rRNA (Figure 5C,D). This functionally important helix is involved in the formation of the binding pocket for A-site-bound tRNA (22,23) and translocation (24). Methylated G1207 is involved in the base-pair formation with C1051. Neighboring nucleotide C1054 contact anticodon of the tRNA that is bound to the A-site directly (Figure 5D). Despite location in the very functionally important region of the 30S subunit, m2G1207 is not involved in the contact with tRNA (Figure 5B). Methyl group should be located in the minor groove of the helix 34 and it is not interacting with any functional group of the ribosome. We couldnot rule out that it can interact with some ligands that binds the A-site, such as IF1, EF-G, class I termination factors, tmRNA or others. Substitution of the G1207 to A is viable and does not have any phenotype (19) despite the fact that C1051-G1207 pair would be substituted by C-A opposition. Transversions of G1207 to both pyrimidines are lethal (19) perhaps due to alternative pairing of 1053GCA1055 with 1207C(U)GU1205 destroying secondary structure of the helix 34. It could be hypothesized that methylation of G1207 may be necessary to freeze secondary structure in the functional conformation, perhaps avoiding formation of the base pairs 1054CA1055 with 1206GU1205.
m2G 1835 OF THE 23S rRNA
Nucleotide 1835 is located in the four-way junction of the helices 67, 68, 69 and 71 of 23S rRNA close to subunit interface (Figure 5E,F). This residue is conserved among all kingdoms of life (18). Although not in direct contact with the small subunit, m2G1835 is surrounded by intersubunit bridges B2a–c, B3 and B7a (25–27). Helix 69 was cross-linked to the 16S rRNA (28). A substantial fraction of nucleotides located in helices 68 and 69 are protected from either hydroxyl radical cleavage or base-specific chemical modification upon subunit association (29). Among bridges surrounding m2G1835 the most conserved is B2a, contributing also to A- and P-site tRNA binding. Mutations of nucleotides C1914 and A1916 of this bridge lead to increase in translational error rates (30). Other mutations within helix 69 severely affected cell growth and translation (31). Methylation of nucleotides A1912 and A1918 by DMS in modification-interference experiments prevented subunit association (32).
23S rRNA area encompassing nucleotides 1835–1962 is one of the most heavily modified in the ribosome, it contains seven modified nucleotides. Among them are three pseudouridine residues, whose formation is vital for the cell (33). On the opposite side, in the 30S ribosomal subunit there is also a cluster of modified nucleotides. The loop of the helix 45 contains five methyl groups attached to three modified nucleotides, two of which are dimethylated adenosine residues, 1518 and 1519. Lack of modification in either nucleotide or mutation of A1519, results in the resistance to antibiotic kasugamicin (34,35). The third modified residue, m2G1516, is of unknown importance and function. The enzyme responsible for the modification has not been identified, although it was suggested to be encoded by the ybiN gene (8).
Lack of G1835 methylation in rlmG(ygjO) knockout strain doesnot lead to significant growth retardation at the optimal growth conditions (10). However, in the poor medium and at elevated temperature, the rlmG(ygjO) knockout strain has significantly decreased fitness (10). The normal subunit association is unlikely to be affected by the lack of G1835 methylation. However, many stress-related factors are known to bind subunit interface at non-optimal conditions (36,37). We can hypothesize that m2G1835 is involved in the regulation of subunit joining at stress conditions.
m2G 2445 OF THE 23S rRNA
Methylated residue G2445 is part of the helix 74 of the 23S rRNA (Figure 5G,H). This helix is directly linked to the peptidyltransferase circle in 23S rRNA secondary structure (18). Several 23S rRNA nucleotides are proximal to m2G2445 in the 50S subunit. First of all, C2065 forms a base pair with m2G2445. Both residues are absolutely conserved among all kingdoms of life (18). Only in some mitochondria U–G and U–A variants of this base pair are tolerated. Lack of G2445 methylation as a consequence of deletion of the rlmL(ycbY) gene causes significant growth retardation, most prominent among all ribosomal guanine-(N2)-methyltransferases yet tested. Growth disadvantage of the rlmL(ycbY) deletion strain is potentiated in the minimal media (11). Since G2445 is monomethylated at the exocyclic amino group, it leaves one hydrogen atom to establish a Watson–Crick base pair. If the pairing with C2065 is maintained, methyl group should be located in the minor groove of the helix, in which it contacts G830 and dU2449 nucleotides (27). Both G830 and dU2449 could potentially form a base triple complex (Figure 5H) forcing C2065–m2G2445 pair. Methyl group prevents formation of the triple and cause G830 and dU2449 to move out of the C2065–m2G2445 plane (Figure 5H). We can hypothesize that methylation of G2445 is necessary to fix the structure of 23S rRNA region proximal to the peptidyltransferase center in conformation, optimal for the function. It can hardly be imagined that methyl group contacts any ribosomal ligand, since m2G2445 is packed in the core of the large subunit. It is also unlikely that methylation of G2445 is necessary for the large ribosomal subunit assembly because no temperature dependence of the cell fitness on the methylation of G2445 was observed (11).
CONCLUSIONS
Up to now four out five ribosomal guanine-(N2)-methyltransferases have been identified. All enzymes that form m2G in rRNA have highly similar architecture of the AdoMet-binding site. According to the ribosomal substrate specificity, they could be divided into two groups. The relatively large enzymes RlmG and RlmL possessing an additional RNA-binding domain act on naked ribosomal RNA or early assembly intermediates in the cell. The smaller RsmC and RsmD methyltransferases utilize assembled small subunit or its late assembly intermediates as a substrate. It is likely that the latter class of proteins uses the decoding cleft of the small subunit as the binding site.
On the basis of the analysis of the ribosome structure, we can propose that m2G-modified residues could be essential for stabilization of correct rRNA fine structure at the functionally important sites by preventing the formation of non-correct interactions. Indeed, m2G966 is involved in fine-tuning of tRNA binding to the P-site. Methylated G1207 might either stabilize the secondary structure of the helix 34 of the 16S rRNA or be involved in interaction with some ribosomal ligands. Perhaps, methylation of G1835 located at the subunit interface is needed at the stress conditions for regulation of subunit joining. The likely role of the 23S rRNA G2445 methylation is to freeze an active conformation of the peptidyltransferase center by preventing of the involvement of C2065–m2G2445 pair in base triple interactions.
A progress in identification of the ribosomal methyltransferase genes makes it highly probable that all the genes encoding such enzymes will be identified in several years. However, the major work on determination of the function of the ribosomal RNA modifications is only in the beginning.
Supplementary Material
[Supplementary Material]
ACKNOWLEDGEMENTS
This work was supported by grants from Howard Hughes Medical Institute, Russian Foundation for Basic Research and Leading Scientific Schools. Funding to pay the Open Access publication charge was provided by Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Andersen NM, Douthwaite S. YebU is a m5C methyltransferase specific for 16S rRNA nucleotide 1407. J. Mol. Biol. 2006;359:777–786. doi: 10.1016/j.jmb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Mueller F, Brimacombe R. A new model for the three-dimensional folding of Escherichia coli 16S Ribosomal RNA. I. Fitting the RNA to a 3D electron microscopic map at 20 Å. J. Mol. Biol. 1997;271:524–544. doi: 10.1006/jmbi.1997.1210. [DOI] [PubMed] [Google Scholar]
- 3.Cundliffe E. Ribosomal modification and resistance in antibiotic-producing organisms. Biochem. Soc. Symp. 1987;53:1–8. [PubMed] [Google Scholar]
- 4.Maravic G. Macrolide resistance based on the Erm-mediated rRNA methylation. Curr. Drug Targets Infect. Disord. 2004;4:193–202. doi: 10.2174/1568005043340777. [DOI] [PubMed] [Google Scholar]
- 5.Del Campo M, Kaya Y, Ofengand J. Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA. 2001;7:1603–1615. [PMC free article] [PubMed] [Google Scholar]
- 6.Tscherne JS, Nurse K, Popienick P, Ofengand J. Purification, cloning, and characterization of the 16S RNA m2G1207 methyltransferase from Escherichia coli. J. Biol. Chem. 1999;274:924–929. doi: 10.1074/jbc.274.2.924. [DOI] [PubMed] [Google Scholar]
- 7.Bujnicki JM. Phylogenomic analysis of 16S rRNA:(guanine-N2) methyltransferases suggests new family members and reveals highly conserved motifs and a domain structure similar to other nucleic acid amino-methyltransferases. FASEB J. 2000;14:2365–2368. doi: 10.1096/fj.00-0076com. [DOI] [PubMed] [Google Scholar]
- 8.Bujnicki JM, Rychlewski L. RNA:(guanine-N2) methyltransferases RsmC/RsmD and their homologs revisited—bioinformatic analysis and prediction of the active site based on the uncharacterized Mj0882 protein structure. BMC Bioinformatics. 2002;3:10. doi: 10.1186/1471-2105-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lesnyak DV, Osipiuk J, Skarina T, Sergiev PV, Bogdanov AA, Edwards A, Savchenko A, Joachimiak A, Dontsova OA. Methyltransferase that modifies guanine 966 of the 16S rRNA: Functional identification and tertiary structure. J. Biol. Chem. 2007;282:5880–5887. doi: 10.1074/jbc.M608214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sergiev PV, Lesnyak DV, Bogdanov AA, Dontsova OA. Identification of Escherichia coli m(2)G methyltransferases: II. The ygjO gene encodes a methyltransferase specific for G1835 of the 23S rRNA. J. Mol. Biol. 2006;364:26–31. doi: 10.1016/j.jmb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Lesnyak DV, Sergiev PV, Bogdanov AA, Dontsova OA. Identification of Escherichia coli m(2)G methyltransferases: I. The ycbY gene encodes a methyltransferase specific for G2445 of the 23S rRNA. J. Mol. Biol. 2006;364:20–25. doi: 10.1016/j.jmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Huang L, Hung L, Odell M, Yokota H, Kim R, Kim SH. Structure-based experimental confirmation of biochemical function to a methyltransferase, MJ0882, from hyperthermophile Methanococcus jannaschii. J. Struct. Funct. Genomics. 2002;2:121–127. doi: 10.1023/a:1021279113558. [DOI] [PubMed] [Google Scholar]
- 13.Weitzmann C, Tumminia SJ, Boublik M, Ofengand J. A paradigm for local conformational control of function in the ribosome: binding of ribosomal protein S19 to Escherichia coli 16S rRNA in the presence of S7 is required for methylation of m2G966 and blocks methylation of m5C967 by their respective methyltransferases. Nucleic Acids Res. 1991;19:7089–7095. doi: 10.1093/nar/19.25.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 15.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 16.Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 17.Selmer M, Dunham CM, Murphy FV, IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 18.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:15. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jemiolo DK, Taurence JS, Giese S. Mutations in 16S rRNA in Escherichia coli at methyl-modified sites: G966, C967, and G1207. Nucleic Acids Res. 1991;19:4259–4265. doi: 10.1093/nar/19.15.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalak JA, Bruenger E, Crain PF, McCloskey JA. Identities and phylogenetic comparisons of posttranscriptional modifications in 16S ribosomal RNA from Haloferax volcanii. J. Biol. Chem. 2000;275:24484–24489. doi: 10.1074/jbc.M002153200. [DOI] [PubMed] [Google Scholar]
- 21.Youvan DC, Hearst JE. A sequence from Drosophila melanogaster 18S rRNA bearing the conserved hypermodified nucleoside am psi: analysis by reverse transcription and high-performance liquid chromatography. Nucl. Acids Res. 1981;9:1723–1741. doi: 10.1093/nar/9.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sergiev PV, Lavrik IN, Dokudovskaya SS, Dontsova OA, Bogdanov AA. Structure of the decoding center of the ribosome. Biochemistry (Mosc.) 1998;63:963–976. [PubMed] [Google Scholar]
- 23.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 24.Kubarenko A, Sergiev P, Wintermeyer W, Dontsova O, Rodnina MV. Involvement of helix 34 of 16S rRNA in decoding and translocation on the ribosome. J. Biol. Chem. 2006;281:35235–35244. doi: 10.1074/jbc.M608060200. [DOI] [PubMed] [Google Scholar]
- 25.Gabashvili IS, Agrawal RK, Spahn CMT, Grassucci RA, Svergun DI, Frank J, Penczek P. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 26.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 27.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Doudna Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell P, Osswald M, Brimacombe R. Identification of intermolecular cross-links at the subunit interface of the E. coli ribosome. Biochemistry. 1992;31:3004–3011. doi: 10.1021/bi00126a023. [DOI] [PubMed] [Google Scholar]
- 29.Merryman C, Moazed D, Daubresse G, Noller HF. Nucleotides in 23S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 1999;285:107–113. doi: 10.1006/jmbi.1998.2243. [DOI] [PubMed] [Google Scholar]
- 30.O'Connor M, Dahlberg AE. The involvement of two distinct regions of 23S ribosomal RNA in tRNA selection. J. Mol. Biol. 1995;254:838–847. doi: 10.1006/jmbi.1995.0659. [DOI] [PubMed] [Google Scholar]
- 31.Liiv A, Karitkina D, Maiväli Ü, Remme J. Analysis of the function of E. coli 23S rRNA helix-loop 69 by mutagenesis. BMC Mol. Biol. 2005;6:18. doi: 10.1186/1471-2199-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maivali U, Remme J. Definition of bases in 23S rRNA essential for ribosomal subunit association. RNA. 2004;10:600–604. doi: 10.1261/rna.5220504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raychaudhuri S, Conrad J, Hall BG, Ofengand J. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA. 1998;4:1407–1417. doi: 10.1017/s1355838298981146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sparling PF. Kasugamycin resistance: 30S ribosomal mutation with an unusual location on the Escherichia coli chromosome. Science. 1970;167:56–58. doi: 10.1126/science.167.3914.56. [DOI] [PubMed] [Google Scholar]
- 35.Vila-Sanjurjo A, Squires CL, Dahlberg AE. Isolation of kasugamycin resistant mutants in the 16S ribosomal RNA of Escherichia coli. J. Mol. Biol. 1999;293:1–8. doi: 10.1006/jmbi.1999.3160. [DOI] [PubMed] [Google Scholar]
- 36.Agafonov DE, Kolb VA, Spirin AS. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep. 2001;2:399–402. doi: 10.1093/embo-reports/kve091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida H, Maki Y, Kato H, Fujisawa H, Izutsu K, Wada C, Wada A. The ribosome modulation factor (RMF) binding site on the 100S ribosome of Escherichia coli. J. Biochem. (Tokyo) 2002;132:983–989. doi: 10.1093/oxfordjournals.jbchem.a003313. [DOI] [PubMed] [Google Scholar]
- 38.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32(W):327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]