Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells (original) (raw)
Abstract
Virus infection triggers IFN immune defenses in infected cells in part through viral nucleic acid interactions, but the pathways by which dsDNA and DNA viruses trigger innate defenses are only partially understood. Here we present evidence that both retinoic acid-induced gene I (RIG-I) and mitochondrial antiviral signaling protein (MAVS) are required for dsDNA-induced IFN-β promoter activation in a human hepatoma cell line (Huh-7), and that activation is efficiently blocked by the hepatitis C virus NS3/4A protease, which is known to block dsRNA signaling by cleaving MAVS. These findings suggest that dsDNA and dsRNA share a common pathway to trigger the innate antiviral defense response in human cells, although dsDNA appears to trigger that pathway upstream of the dsRNA-interacting protein RIG-I.
Keywords: IFN-β, DNA virus, hepatitis C virus, retinoic acid-induced gene I
The host innate immune system senses nonself entities through specific molecular pattern recognition (1). Nonself nucleic acids are recognized by two types of receptors. Toll-like receptors (TLRs) are localized on the cell surface or in endosomes. dsRNA is recognized by TLR-3, whereas ssRNA is recognized by TLR-7 and -8. TLR-9, a sensor of foreign DNA, recognizes certain CpG oligodeoxynucleotides (2). However, only a subset of cells that activate type I IFN in response to pathogen invasion do so through TLRs (1). Recent studies have shown that intracellular dsRNA can be recognized by another group of receptors that are TLR-3-independent (3, 4). Intracellular dsRNA can be specifically recognized by either retinoic acid-induced gene I (RIG-I) (3) or melanoma differentiation-associated gene 5 (MDA5) (4). Both RIG-I and MDA5 contain DExD/H-box helicase domains and caspase recruitment domains (CARDs). After dsRNA binding occurs, RIG-I and MDA5 use their tandem CARD domains to interact with the CARD domains of mitochondrial antiviral signaling protein (MAVS) (also known as IPS-1, Cardif, or VISA) (5).
Notably, both the TLR-3 and the RIG-I/MDA5 signaling pathways induce type I IFN through the activation of IFN regulatory factor 3 (IRF-3) or IRF-7 (6). IRF-3 and -7 normally reside in the cytoplasm in an inactive state. Phosphorylation by the kinases Tank-binding kinase (TBK)-1- or I-κB kinase-ε (IKK-ε) triggers IRF-3 and -7 nuclear translocation and transcription of type I IFN genes (1, 5). In addition, both signaling pathways activate NF-κB and the expression of inflammatory cytokines (1, 3). As a countermeasure, many viruses have evolved strategies to inhibit the innate signaling events leading to IFN production. For example, the hepatitis C virus (HCV) NS3/4A protease blocks the RIG-I-mediated signaling pathway by cleaving the MAVS protein and blocking downstream IFN-β gene expression (7–10).
TLR-9, the only known primary sensor of foreign DNA, recognizes unmethylated CpG DNA (11). Accumulating evidence, however, has suggested that DNA can also be recognized by a TLR-9 independent receptor (12–14). For example, DNA derived from either pathogens or the host activates the innate immune response when transfected into the cytoplasm (12, 13, 15), and this activation depends on the double-stranded structure of DNA (16). Secondly, DNase II-deficient macrophages that cannot degrade DNA from phagocytosed apoptotic cells produce IFN-β independently of TLR-9 (14, 17). Furthermore, microarray analysis reveals an overlapping but unique gene expression profile activated by intracellular (cytosolic) DNA compared with the TLR-9-mediated response (13).
Recent reports have shown that IFN-β promoter activation by cytosolic dsDNA requires the transcription factor IRF-3 and the kinases TBK-1/IKK-ε, and is independent of TLRs, NOD proteins, and RIP2 (12, 13, 15). Additionally, cytosolic dsDNA-induced IFN-β production in human HEK293 cells has been partially suppressed by MAVS-specific siRNA (12), suggesting that MAVS may mediate dsDNA-signaling in human cells. In contrast, dsDNA-induced IFN-β production is normal in RIG-I-deficient mouse embryo fibroblasts (MEFs) (12) and in MAVS-deficient MEFs (18, 19), indicating that dsDNA signaling is both RIG-I- and MAVS-independent in mice. Thus, it has been hypothesized that cytosolic DNA is recognized by an unknown sensor that is different from dsRNA sensors and transduces the DNA signal through the activation of TBK-1/IKK-ε (18, 19).
In this report, we used the Huh-7 human hepatoma cell line to demonstrate that dsDNA is a potent inducer of IFN-β and IFN-stimulated gene expression. In contrast to the results obtained in murine systems, we found that both RIG-I and MAVS are essential for the cytosolic dsDNA-signaling pathway in human cells. Collectively, these results demonstrate that a common signaling pathway is triggered by dsDNA and dsRNA in human cells, and they imply that the dsDNA-sensing machinery is different in mice and humans.
Results
Activation of IFN-β Expression by Cytosolic dsDNA in Huh-7 Cells.
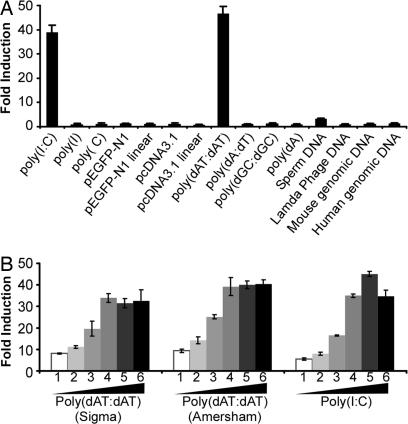
To determine whether cytosolic dsDNA can activate IFN-β production in Huh-7 cells, cells were first transfected with the IFN-β promoter luciferase reporter. Thirty-six hours after transfection, cells were either mock transfected or transfected with various DNA stimuli (1.0 μg/ml) by using Lipofectamine 2000. In parallel, a synthetic form of dsRNA, poly(I):poly(C) [poly(I:C)], was transfected as a positive control. As reported (10), dsRNA but not ssRNA induced IFN-β expression in Huh-7 cells (Fig. 1A). Importantly, a synthetic form of dsDNA, poly(dA-dT):poly(dA-dT) [poly(dAT:dAT)], was also able to induce IFN-β promoter activation (Fig. 1A) and IFN-stimulated gene expression [see supporting information (SI) Fig. 6_A_] in the cells. In addition, intracellular poly(dAT:dAT) was able to efficiently induce NF-κb responsive genes, e.g., CCl5, CxCL9, CxCL10, and ICAM-1 (SI Fig. 6_B_). Interestingly, other dsDNAs, including mouse and human genomic DNA, sperm DNA, plasmid DNA, and other synthetic ssDNA or dsDNA, failed to or very weakly activated the IFN-β promoter (Fig. 1A). Similar results were obtained when these DNAs were transfected into Huh-7 cells at a higher concentration (10.0 μg/ml) (data not shown).
Fig. 1.
Cytosolic dsDNA activates the IFN-β promoter in Huh-7 cells. (A) Huh-7 cells were first cotransfected with the IFN-β promoter luciferase reporter plasmid, an internal control plasmid pRL-TK, and a carrier plasmid pcDNA3.1. At 36 h after transfection, cells were either mock-transfected or transfected with various DNA or RNA stimuli (1.0 μg/ml) using Lipofectamine 2000. Cells were subjected to dual luciferase assay 16 h after transfection. The results are expressed as fold induction of IFN-β promoter activity relative to the basal level. (B) Dose titration of dsDNA and dsRNA to induce IFN-β promoter activation. The experimental conditions were the same as in A except that the indicated amount of dsDNA or dsRNA was transfected. (1-6: 0.125, 0.25, 0.5, 1.0, 2.0, and 4.0 μg/ml).
To confirm the specific induction of IFN-β promoter activation by intracellular dsDNA poly(dAT:dAT), three additional experiments were carried out. First, the poly(dAT:dAT) purchased from a different company (Sigma, St. Louis, MO) was tested, and the results shown in Fig. 1B indicate that the two dsDNAs activate the IFN-β promoter equally well. Dose titration of the two dsDNAs and dsRNA clearly shows that the poly(dAT:dAT) is at least as efficient as poly(I:C) in Huh-7 cells (Fig. 1B). Second, to rule out the possibility that these results reflected the presence of trace amounts of dsRNA contaminants in the dsDNA preparations, we showed that poly(dAT:dAT) is fully active after RNase A digestion but not after DNase I digestion (SI Fig. 6_C_). Finally, although transfected poly(dAT:dAT) efficiently induced IFN-β or IFN-stimulated gene expression in Huh-7 cells, it failed to do so when it was simply added to the culture medium (SI Fig. 6_A_). Taken together, these results suggest that, like dsRNA, intracellular (cytosolic) dsDNA, in particular poly(dAT:dAT), can be sensed in Huh-7 cells, where it efficiently triggers the host innate immune response to induce IFN-β.
dsDNA-Induced IFN-β Expression Depends on MAVS.
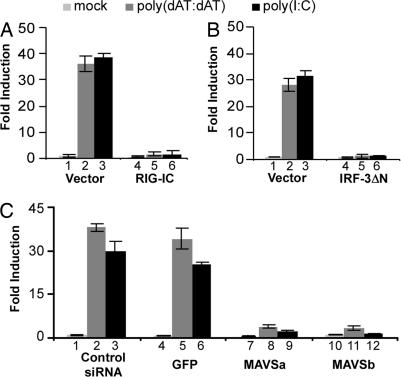
Previous studies in mouse cells have shown that dsDNA-induced IFN-β expression depends on IRF-3 and TBK-1/IKK-ε but not MAVS or RIG-I (12, 13, 18, 19). To further characterize dsDNA-induced signaling, we first cotransfected Huh-7 cells with a construct that expresses either a dominant-negative form of IRF-3 (IRF-3 ΔN) or a dominant-negative form of RIG-I (RIG-IC) together with the IFN-β promoter luciferase reporter, and we examined them for IFN-β promoter activity with and without transfection of poly(dAT:dAT) or poly(I:C).
As shown in Fig. 2 A and B, consistent with the known requirement of RIG-I and IRF-3 for intracellular dsRNA signaling, expression of either of the dominant-negative mutants RIG-IC or IRF-3 ΔN completely blocked dsRNA-induced IFN-β promoter activation. Similarly, expression of both the RIG-I and the IRF-3 dominant-negative mutants completely blocked dsDNA-induced IFN-β promoter activation (Fig. 2 A and B). The RIG-I results are surprising, because previous results in mouse cells suggested that RIG-I is dispensable for signaling by the dsDNA pathway. Confirming these results, we showed that RIG-IC expression blocks dsDNA signaling in 293T cells as well as in Huh-7 cells (SI Fig. 7).
Fig. 2.
MAVS is essential for dsDNA-induced IFN-β promoter activation. (A and B) RIG-I and IRF-3 dominant-negative mutants block dsDNA-induced IFN-β promoter activation. Huh-7 cells were cotransfected with the IFN-β promoter luciferase reporter and plasmid pRL-TK in the presence of an empty vector or a plasmid expressing RIG-IC or IRF-3 ΔN. Thirty-six hours later, cells were mock-transfected or transfected with poly(dAT:dAT) or poly(I:C). Luciferase activities were measured 16 h after transfection. (C) Knockdown of MAVS blocks dsDNA-induced IFN-β promoter activation. siRNAs were transfected into Huh-7 cells as described in Material and Methods. At 24 h after transfection, cells were transfected with the IFN-β promoter luciferase reporter and subjected to the luciferase reporter assay.
Previous studies, mostly performed in mouse cells, suggest that cytosolic dsDNA has a unique sensor whose downstream signaling events converge with the dsRNA signaling pathway at the level of TBK-1 and IRF-3 (12, 13, 18, 19). The results shown in Fig. 2B indicate that IRF-3 is required for dsDNA signaling, which is further supported by dsDNA-induced IRF-3 nuclear accumulation, a hallmark of its activation (SI Fig. 8). However, the blockade of dsDNA signaling by RIG-IC indicates that RIG-I and, perhaps other upstream signaling components, e.g., MAVS, could also be important for dsDNA signaling in human cell lines. To examine this possibility, we asked whether MAVS is required for dsDNA signaling by using siRNAs to specifically inhibit MAVS gene expression in Huh-7 cells. Compared with a negative-control siRNA or unrelated GFP siRNA, two independent MAVS-specific siRNAs efficiently suppressed MAVS mRNA by ≈85% (SI Fig. 9_A_). As expected, transfection of either MAVS siRNA into Huh-7 cells inhibited dsRNA-induced IFN-β promoter activation by ≈10-fold, confirming the essential role of MAVS in the dsRNA signaling pathway (Fig. 2C, lanes 9 and 12). Importantly, transfection of MAVS siRNA inhibited dsDNA-induced IFN-β promoter activation to a similar degree (Fig. 2C, lanes 8 and 11). Confirming this observation, both of the MAVS siRNAs efficiently blocked IFN-β promoter activation induced by a constitutively active form of RIG-I (RIG-IN) or MAVS but not by a constitutively active form of IRF-3 (IRF-3 5D), which acts downstream of MAVS (SI Fig. 9_C_). These results strongly suggest that MAVS is essential for dsDNA signaling in human cells.
dsDNA-Induced IFN-β Expression Is Blocked by the HCV NS3/4A Protease.
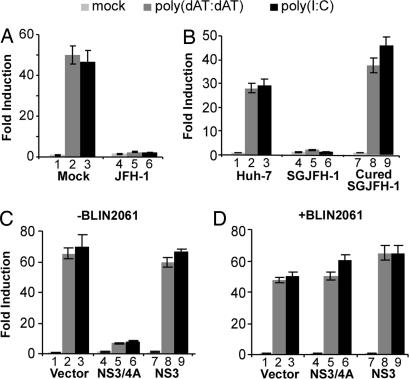
We and others have recently reported that the HCV NS3/4A protease prevents IFN-β promoter activation by proteolytically cleaving MAVS (7–10). To further validate the essential role of MAVS in the dsDNA signaling pathway, we examined whether HCV infection blocks this pathway.
Huh-7 cells were infected by the HCV JFH-1 virus. At day 4 after infection, when >90% of the cells were infected (not shown), cells were transfected with the IFN-β promoter reporter construct. Thirty-six hours later, cells were then either mock-transfected or transfected with poly(dAT:dAT) or poly(I:C). HCV infection did not decrease cell transfection efficiency, because HCV-infected cells displayed comparable levels of activity of the control plasmid (pRL-TK) compared with mock-infected cells (data not shown). As shown in Fig. 3A, the IFN-β promoter was activated ≈40-fold in mock-infected cells transfected with either poly(dAT:dAT) or poly(I:C) (Fig. 3A, lanes 2 and 3), whereas it was not activated by either stimulus in HCV-infected cells (Fig. 3A, lanes 5 and 6), suggesting that HCV infection blocks the dsDNA signaling pathway. Moreover, the dsDNA signaling pathway was blocked in JFH-1 subgenomic replicon cells (Fig. 3B, lane 4–6) and in cells containing a replicon of a different HCV genotype (Con-1) (data not shown) but not in cured replicon cells (Fig. 3B, lanes 7–9), further indicated that the dsDNA signaling pathway is blocked by HCV.
Fig. 3.
HCV NS3/4A blocks dsDNA-induced IFN-β promoter activation. (A) HCV infection blocks dsDNA-induced IFN-β promoter activation. Huh-7 cells were either mock-infected or infected with JFH-1 at a multiplicity of infection of 1. On day 4 after infection, cells were transfected with the IFN-β promoter luciferase reporter and subjected to the luciferase reporter assay. (B) dsDNA-induced IFN-β promoter activation is blocked in the HCV replicon cells. Parental Huh-7 cells, JFH-1 subgenomic replicon cells (SGJFH-1) and cured replicon cells were transfected with the IFN-β promoter luciferase reporter and subjected to the luciferase reporter assay. (C) HCV NS3/4A protease blocks dsDNA-induced IFN-β promoter activation. Huh-7 cells were cotransfected with the IFN-β promoter luciferase reporter and plasmid pRL-TK in the presence of an empty vector or a plasmid expressing either NS3 or NS3/4A. 36 h later, the IFN-β luciferase activity induced by poly(dAT:dAT) or poly(I:C) was assayed as described in Materials and Methods. (D) BILN2061 prevents the blocking activity of NS3/4A. The experimental conditions were the same as in C, except that the protease inhibitor BILN2061 was added to the cells 1 h before plasmid transfection at a final concentration of 5 μM.
To determine whether the blockade is mediated by viral NS3/4A protease, JFH-1 NS3/4A was overexpressed in Huh-7 cells, which were then transfected with either poly(dAT:dAT) or poly(I:C). The results shown in Fig. 3C clearly demonstrate that the HCV NS3/4A protein could efficiently block the dsDNA signaling pathway. However, NS3 alone had no effect, suggesting that viral protease activity, which depends on NS3-NS4A interactions (20), is critical for the inhibitory effect. Indeed, addition of the specific NS3/4A protease inhibitor BILN2061 completely blocked the inhibitory effect of NS3/4A (Fig. 3D), confirming that HCV interrupts the dsDNA signaling pathway by virtue of its NS3/4A protease activity. Because HCV NS3/4A is known to cleave MAVS (7–10), these results provide further evidence that MAVS is essential for dsDNA signaling.
RIG-I Is Required for dsDNA-Induced IFN-β Expression in Huh-7 Cells.
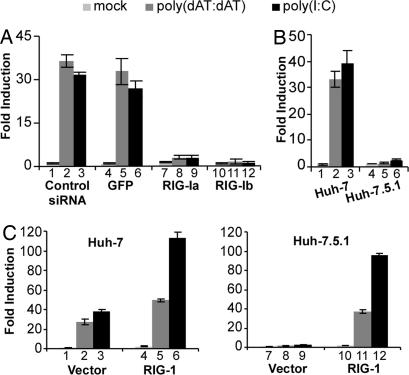
The inhibitory activity of RIG-IC and the essential role of MAVS in the dsDNA signaling pathway suggest that RIG-I could be important for cytosolic dsDNA-dependent signaling in human cell lines. To elucidate the role of RIG-I in dsDNA signaling, two independent siRNAs targeting RIG-I (RIG-Ia was used in ref. 21) were used to suppress RIG-I expression in Huh-7 cells. Compared with the negative control siRNA and an unrelated GFP siRNA, both siRNAs efficiently reduced RIG-I mRNA levels by 70–80% (SI Fig. 9_B_). Importantly, transfection of either of the RIG-I siRNAs into Huh-7 cells inhibited dsDNA-induced IFN-β promoter activation ≈10-fold, similar to their effect on dsRNA-induced signaling (Fig. 4A). These results indicate that RIG-I is essential for the dsDNA signaling pathway.
Fig. 4.
RIG-I is essential for the dsDNA-induced IFN-β promoter activation. (A) Knockdown of RIG-I blocks dsDNA-induced IFN-β promoter activation. siRNAs were transfected into Huh-7 cells as described in Materials and Methods. At 24 h after transfection, cells were transfected with the IFN-β promoter luciferase reporter and subjected to the luciferase reporter assay. (B) dsDNA-induced IFN-β promoter activation is absent in Huh-7.5.1 cells. Huh-7 and Huh-7.5.1 cells were transfected with the IFN-β promoter luciferase reporter and the luciferase activity induced by poly(dAT:dAT) or poly(I:C) was assayed as described in Materials and Methods. (C) Wild-type RIG-I restores dsDNA-induced IFN-β promoter activation in Huh-7.5.1 cells. Huh-7 (Left) or Huh-7.5.1 (Right) cells were cotransfected with the IFN-β promoter luciferase reporter and plasmid pRL-TK in the presence of an empty vector or a plasmid expressing wild-type RIG-I. The IFN-β luciferase activity induced by poly(dAT:dAT) or poly(I:C) was assayed as in B.
To confirm that RIG-I is required for dsDNA signaling, we took advantage of the Huh-7-derived cell line Huh-7.5.1 (22) that contains an inactivating point mutation in RIG-I (23) and therefore is unresponsive to dsRNA (Fig. 4B, compare lane 3 with lane 6). However, the IFN-β promoter is activated equally well in Huh-7 and Huh-7.5.1 cells by expression of a constitutively active form of RIG-I (RIG-IN), as well as by wild-type MAVS and IRF-3 5D, suggesting that the dsRNA signaling pathway downstream of RIG-I is fully functional in these cells (SI Fig. 10_A_). Strikingly, transfected dsDNA poly(dAT:dAT) failed to induce IFN-β promoter activation in Huh-7.5.1 cells (Fig. 4B, compare lane 2 with lane 5), further demonstrating that RIG-I plays an essential role in the dsDNA signaling pathway.
To confirm that the dsDNA signaling deficiency in Huh-7.5.1 cells reflects their inactivating RIG-I mutation, we asked whether wild-type RIG-I could restore dsDNA signaling in these cells. As shown in Fig. 4C, RIG-I overexpression slightly enhanced poly(I:C) or poly(dAT:dAT)-induced IFN-β promoter activation (Fig. 4C Left), but it enabled virtually unresponsive Huh-7.5.1 cells to become highly responsive to transfected poly(I:C) or poly(dAT:dAT) (Fig. 4C Right). Transfection of poly(I:C) or poly(dAT:dAT) increased IFN-β promoter activation by 97- and 37-fold, respectively, in Huh-7.5.1 cells overexpressing RIG-I (Fig. 4C, lanes 11 and 12), compared with 2.3- and 1.4-fold induction in Huh-7.5.1 cells transfected with the empty vector (Fig. 4C, lanes 8 and 9) and to 113- and 49-fold induction in Huh-7 cells overexpressing RIG-I (Fig. 4C, lanes 5 and 6). These results demonstrate that overexpression of wild-type RIG-I is sufficient to restore the dsRNA and dsDNA signaling in Huh-7.5.1 cells. In contrast, signaling components downstream of RIG-I, including TBK-1, IKK-ε, and IRF-3 (MAVS was not included because the IFN-β promoter is strongly induced by its overexpression; see SI Fig. 10_A_), failed to restore the responsiveness to dsRNA or dsDNA in Huh-7.5.1 cells, although overexpression of either of them induced IFN-β promoter activity to an equivalent degree in Huh-7.5.1 and Huh-7 cells. (SI Fig. 10 A and B). Collectively, these results demonstrate that RIG-I plays an essential role in the dsDNA signaling pathway.
DNA Viruses Can Activate the dsDNA Signaling Pathway.
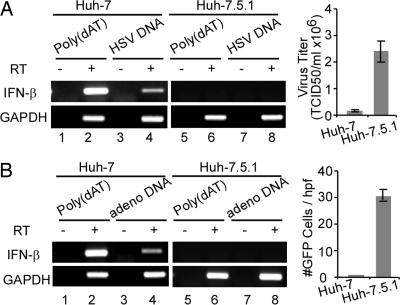
To determine the role of dsDNA signaling in DNA virus infections, Huh-7 and Huh-7.5.1 cells were infected with herpes simplex virus 1 (HSV-1) or adenovirus, as described in Materials and Methods. As shown in Fig. 5, both HSV-1 and adenovirus infection was much more robust in Huh-7.5.1 than in Huh-7 cells. Moreover, we showed that HSV-1 and adenovirus DNA induce IFN-β mRNA production when transfected into Huh-7 cells but not Huh-7.5.1 cells, supporting the notion that both of these DNA viruses have the potential to trigger the dsDNA signaling pathway, and that they do so in a RIG-I-dependent manner.
Fig. 5.
Huh-7.5.1 cells are more permissive to DNA virus infections. (A) HSV-1 infection is more productive in Huh-7.5.1 cells than in Huh-7 cells. (Left) Huh-7 and Huh-7.5.1 cells were transfected with purified HSV-1 DNA at a concentration of 10 μg/ml by using lipofectamine 2000. At 18 h after transfection, cells were harvested and the transcriptional levels of IFN-β and GAPDH were analyzed by using RT-PCR described in Material and Methods. RT reactions without reverse transcriptase were used for DNA contamination control. (Right) Huh-7 and Huh-7.5.1 cells were infected with HSV-1 at multiplicity of infection of 0.05. At 24 h after infection, HSV-1-infected cells were harvested and virus titers were determined in Vero cells. (B) Adenovirus replicates more efficiently in Huh-7.5.1 cells. The DNA transfection and adenovirus infection were done similarly as described in A, except that adenovirus infection was measured by the number of GFP-expressing cells.
Discussion
The results of this study provide evidence that cytosolic dsDNA is a potent inducer of IFN-β expression in human hepatoma Huh-7 cells, and that this activation requires both the intracellular dsRNA sensor RIG-I and its adaptor molecule MAVS, in addition to TBK-1, IKK-ε, and IRF-3, which were previously known to be required for dsDNA signaling (12, 13, 15). Our results thus provide a previously undescribed finding that RIG-I is required for dsDNA signaling in human cells, and they support and extend previous findings by Ishii et al. (12, 18, 19) that MAVS is required for dsDNA signaling in human cells. Notably, siRNA-mediated suppression of MAVS expression as well as the HCV NS3/4A protease, which cleaves and inactivates MAVS, blocked dsDNA-induced signaling. Furthermore, RIG-I, an intracellular dsRNA sensor, was shown to be essential for dsDNA signaling as well. It is noteworthy that a single point mutation in RIG-I in Huh-7.5.1 cells that renders RIG-I incapable of signaling dsRNA also inhibits cell responsiveness to dsDNA. In particular, overexpression of wild-type RIG-I in Huh-7.5.1 cells restored the dsDNA signaling pathway. These findings demonstrate that the dsDNA- and dsRNA-induced innate immune signaling pathways share more components in human cells than originally believed and imply the existence of a mouse-specific dsDNA sensing machinery.
The different roles of RIG-I and MAVS in the human and murine dsDNA signaling pathway are particularly intriguing. The results presented here clearly demonstrate that both RIG-I and MAVS are essential for the dsDNA signaling pathway in human cells. However, convincing evidence from experiments using RIG-I- and MAVS-deficient MEFs demonstrated that neither of these molecules is essential for the dsDNA signaling pathway in mice (12, 18, 19). It is unlikely that these differences are because of the dsDNA reagent poly(dAT:dAT), because it was obtained from the same source in all studies. An alternative explanation for these findings is that the roles of RIG-I and MAVS in the dsDNA signaling pathway are species-specific. In support of this, distinct roles for MAVS in mouse and human cells have also been observed by Ishii et al. and Kumar et al. (12, 18). Moreover, although the type I IFN response to bacteria or DNA virus infection is independent of MAVS in MEFs (18, 19, 24), it is essential in human lung epithelial cells (24). Further studies are needed to validate this hypothesis.
The requirement for RIG-I in dsDNA signaling is supported by evidence obtained using a dominant-negative mutant, siRNAs, and a cell line (Huh-7.5.1) with an inactivating point mutation in RIG-I (23). Importantly, we showed that HSV-1 and adenovirus DNA induce IFN-β mRNA production when they are transfected into Huh-7 but not Huh-7.5.1 cells (Fig. 5), supporting the notion that viral dsDNA has the potential to trigger the dsDNA signaling pathway, and that it does so in a RIG-I-dependent manner. Interestingly, the single point mutation in RIG-I renders Huh-7.5.1 cells unresponsive to both dsRNA and dsDNA, suggesting that both ligands may be recognized in a very similar manner. Indeed, overexpression of wild-type RIG-I in Huh-7.5.1 cells completely restored both the dsRNA and the dsDNA signaling pathway, suggesting that RIG-I may act as a sensor of both dsRNA and dsDNA. To test this hypothesis directly, we performed pulldown experiments with poly(I:C)- and poly(dAT:dAT)-conjugated beads (see SI Text) to determine whether RIG-I can bind dsDNA similar to its known ability to bind to dsRNA (3, 23). Interestingly, RIG-I was efficiently pulled-down by poly(I:C)- but not by the poly(dAT:dAT)-conjugated beads (not shown). Furthermore, RIG-I binding to poly(I:C)-conjugated beads was not reduced by a 100-fold excess of poly(dAT:dAT). These results suggest that, although RIG-I mediates the dsDNA signaling response, it probably does not directly bind dsDNA itself. This suggests the existence of an upstream signaling molecule that senses dsDNA and signals through RIG-I to induce IFN-β expression.
Another interesting observation in this study was that the HCV NS3/4A protease blocks dsDNA-induced IFN-β promoter activation in Huh-7 cells. We and others have shown that HCV uses the NS3/4A protease to block dsRNA signaling and, thereby, prevents IFN-β expression (7–10). Whether the ability of NS3/4A to block dsDNA-induced IFN-β expression plays any role in the pathogenesis of HCV infection is currently unknown. Theoretically, it could facilitate the establishment and persistence of viral infection by cleaving MAVS when immunostimulatory dsDNA is produced, such as from apoptotic cells. More studies will be necessary to examine this possibility.
Finally, how does the cytosolic dsDNA pathway differentiate between self and nonself dsDNA? Self (i.e., genomic) DNA is normally excluded from the cytosol and therefore inaccessible. However, when DNA virus infection or intracellular bacterial infection occurs, DNA release into the cytosol could be recognized by the innate dsDNA signaling sensor (13). In addition, the innate immune system might be able to differentiate between self and foreign nucleic acids. For example, poly(dAT:dAT) was the strongest DNA inducer of IFN-β gene expression compared with other forms of dsDNA, suggesting that a certain DNA structure is preferentially recognized. Recently, one study elegantly demonstrated that the B- but not Z-form of DNA possesses stimulatory activity (12). Moreover, it has been reported that HSV-1 (25) has multiple AT-rich regions in its genome, and certain other pathogens [e.g., Plasmodium falciparum (26)] also contain AT-rich genomes. Interestingly, recent studies indicate that, in bacteria at least, host cells can detect and respond to foreign DNA based on their differences in AT content (27). In addition, poly(dA:dT) induces dsDNA signaling much more efficiently in mouse macrophages than in MEFs (12, 19), suggesting that signaling is probably both species- and cell type-specific. Clearly, self/nonself discrimination is important for the host, because its absence or failure could result in IFN-dependent autoimmunity (14, 17). Further studies are required to provide more insight into the mechanisms of DNA-induced innate immune activation, host defense, and DNA-associated immune disorders.
Materials and Methods
Cell Culture and Reagents.
Huh-7 and Huh-7.5.1 cells were described (22). HEK-derived 293T cells, Vero cells, and HSV-1 (F strain) were obtained from American Type Culture Collection (Bethesda, MD). Adenovirus (Ad5-GFP) was provided by U. Protzer (28). All cells were maintained as described (22). Poly(I:C), polyI, and polyC were purchased from Sigma. Poly(dAT:dAT), poly(dA:dT), poly(dGC:dGC), poly(dA), and sperm DNA were purchased from Amersham Biosciences (Pittsburgh, PA). Plasmid pEGFP-N1 and mouse and human genomic DNA were purchased from Clontech (Mountain View, CA). DNase I and RNase A were purchased from Ambion (Austin, TX) and Invitrogen (Carlsbad, CA), respectively.
Plasmids.
The IFN-β promoter-containing plasmid, pIFΔ(-125) lucter, and the expression plasmids pRL-TK, TBK-1, IKK-ε, and MAVS have been described (10). The plasmids of the wild-type IRF-3, the constitutively active form IRF-3(5D), and the dominant-negative form IRF-3 ΔN were gifts from J. Hiscott (McGill University, Montreal, PQ, Canada) (29). The expression plasmids for wild-type RIG-I, its constitutively active mutant (RIG-IN), and the dominant-negative mutant (RIG-IC) were obtained from T. Fujita (3). To construct expression plasmids of HCV NS3 and NS3/4A proteins, DNA fragments encoding the corresponding genes were amplified from the JFH-1 subgenomic replicon construct pSGJFH-1 (30) by PCR and cloned into the expression vector pcDNA3.1 (Invitrogen). The PCR primers used for NS3 and NS3/4A are listed in SI Table 1.
HCV Infection and RNA Analysis.
These experiments were performed as described in ref. 10. Details are provided in SI Text.
DNA Virus Infection and Viral DNA Preparation.
Huh-7 and Huh-7.5.1 cells were seeded at a density of 6 × 105 cells in T25 flasks. After overnight culture, cells were infected with virus at multiplicity of infection 0.05 and 1.0 for HSV-1 and adenovirus, respectively, and incubated at 37°C in complete growth medium. At 24 h postinfection, the virus yield (TCID50/ml) of HSV-1 infection was titrated in Vero cells as described (31). Adenovirus infection was measured by GFP expression. HSV-1 viral DNA was prepared from infected Vero cells, as described (31). Adenovirus virus DNA was extracted from infected 293 cells by modified Hirt's procedure (28). Both viral DNAs were digested by RNase A and purified by standard DNA phenol/chloroform extraction, as described (32).
RNA Interference.
The siRNA duplex composed of 21-bp sense and antisense oligonucleotides was synthesized by Qiagen (Valencia, CA). The sequences of the siRNA oligos used in this study were as follows (only the sense strands are shown): MAVSa (899–917), CCACCUUGAUGCCUGUGAAUU; MAVSb (142–162) UUGCUGAAGACAAGACCUAUA; RIG-Ia (2363–2381), AAUUCAUCAGAGAUAGUCAUU; RIG-Ib: (358–378), and AAGGCUGGUUCCGUGGCUUUU. Control siRNAs (AllStars Negative Control siRNA and GFP-siRNA) were purchased from Qiagen. Huh-7 cells were transfected with 50 nM siRNA by using _Trans_IT-siQUEST (Mirus Bio Corp., Madison, WI) according to the manufacturer's protocol. At 24 h after transfection, cells were subjected to plasmid transfection and reporter assay. Knockdown of targeted genes was verified by RT–quantitative PCR.
Transfection and Reporter Assay.
The plasmid DNA transfection and IFN-β promoter luciferase reporter assay were performed as described (10). In selected experiments, 36 h after transfection, cells were either mock-transfected or transfected with 0.3 μg of various dsDNA or dsRNA stimuli, using Lipofectamine 2000 (Invitrogen). To examine the effect of the protease inhibitor BILN2061 (33) (Boehringer Ingelheim, Quebec, PQ, Canada), cells were cultured with medium containing 5 μM BILN2061 for 1 h before transfection. All experiments were performed at least twice, with at least duplicate samples in each study. Results are presented as mean ± standard deviation of a representative experiment.
Supplementary Material
Supporting Information
Acknowledgments
We thank Dr. Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) for providing the JFH-1 cDNA plasmid, Dr. S. Goodbourn (University of London, London, U.K.) for providing the pIFΔ(-125) lucter plasmid, Dr. J. Hiscott (McGill University, Montreal, PQ, Canada) for the IRF-3 expression constructs, Dr. K. Fitzgerald (University of Massachusetts Medical School, Worcester, MA) for the TBK-1 and IKK-ε expression plasmids, Dr. T. Fujita (Tokyo Metropolitan Institute, Tokyo, Japan) for the RIG-I expression plasmids, and Dr. Z. Chen (University of Texas Southwestern Medical Center, Dallas, TX) for the MAVS expression construct. BILN2061 was provided by Boehringer Ingelheim. We also thank Dr. H. Maier for critical reading of the manuscript and excellent suggestions and the Molecular Biology Service laboratory of The Scripps Research Institute Department of Molecular and Experimental Medicine (supported by the Sam and Rose Stein Endowment Fund) for oligo synthesis and sequencing analysis. This work was supported by National Institutes of Health Grant R01-CA108304 and by a gift from Mr. Clifford Evans. This is manuscript No. 18670-MEM from The Scripps Research Institute.
Abbreviations
HCV
hepatitis C virus
poly(I:C)
poly(I):poly(C)
poly(dAT:dAT)
poly(dA-dT):poly(dA-dT)
IRF-3
IFN-regulatory factor 3
TBK-1
Tank-binding kinase-1
IKK-ε
I-κB kinase-ε
TLR
Toll-like receptor
IRF
IFN regulatory factor
MEF
mouse embryo fibroblast
HSV
herpes simplex virus.
Footnotes
The authors declare no conflict of interest.
References
- 1.Akira S, Uematsu S, Takeuchi O. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. Curr Opin Immunol. 2005;17:338–344. doi: 10.1016/j.coi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 4.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, et al. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 5.Hiscott J, Lin R, Nakhaei P, Paz S. Trends Mol Med. 2006;12:53–56. doi: 10.1016/j.molmed.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Honda K, Taniguchi T. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 7.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 8.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Proc Natl Acad Sci USA. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, et al. Proc Natl Acad Sci USA. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng G, Zhong J, Chisari FV. Proc Natl Acad Sci USA. 2006;103:8499–8504. doi: 10.1073/pnas.0602957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 12.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 13.Stetson DB, Medzhitov R. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 15.Stetson DB, Medzhitov R. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki K, Mori A, Ishii KJ, Saito J, Singer DS, Klinman DM, Krause PR, Kohn LD. Proc Natl Acad Sci USA. 1999;96:2285–2290. doi: 10.1073/pnas.96.5.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S. J Exp Med. 2005;202:1333–1339. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Failla C, Tomei L, De Francesco R. J Virol. 1994;68:3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seth RB, Sun L, Ea CK, Chen ZJ. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Opitz B, Vinzing M, van Laak V, Schmeck B, Heine G, Gunther S, Preissner R, Slevogt H, N'Guessan PD, Eitel J, et al. J Biol Chem. 2006;281:36173–36179. doi: 10.1074/jbc.M604638200. [DOI] [PubMed] [Google Scholar]
- 25.Lee SS, Lehman IR. Proc Natl Acad Sci USA. 1997;94:2838–2842. doi: 10.1073/pnas.94.7.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber JL. Gene. 1987;52:103–109. doi: 10.1016/0378-1119(87)90399-4. [DOI] [PubMed] [Google Scholar]
- 27.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 28.Sprinzl MF, Oberwinkler H, Schaller H, Protzer U. J Virol. 2001;75:5108–5118. doi: 10.1128/JVI.75.11.5108-5118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin R, Heylbroeck C, Pitha PM, Hiscott J. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Cheng G, Yang K, He B. J Virol. 2003;77:10154–10161. doi: 10.1128/JVI.77.18.10154-10161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 33.Thibeault D, Bousquet C, Gingras R, Lagace L, Maurice R, White PW, Lamarre D. J Virol. 2004;78:7352–7359. doi: 10.1128/JVI.78.14.7352-7359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information