A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae (original) (raw)
Abstract
Cryptic unstable transcripts (CUTs) are synthesized from intra- and intergenic regions in Saccharomyces cerevisiae and are rapidly degraded by RNA surveillance pathways, but their function(s) remain(s) elusive. Here, we show that an antisense TY1 CUT, starting within the Ty1 retrotransposon and encompassing the promoter 5′ long terminal repeat (LTR), mediates RNA-dependent gene silencing and represses Ty1 mobility. We show that the Ty1 regulatory RNA is synthesized by RNA polymerase II, polyadenylated, and destabilized by the cytoplasmic 5′ RNA degradation pathway. Moreover, the Ty1 regulatory RNA represses Ty1 transcription and transposition in trans by acting on the de novo transcribed TY1 RNA. Consistent with a transcriptional regulation mechanism, we show that RNA polymerase II occupancy is reduced on the Ty1 chromatin upon silencing, although TBP binding remains unchanged. Furthermore, the Ty1 silencing is partially mediated by histone deacetylation and requires Set1-dependent histone methylation, pointing out an analogy with heterochromatin gene silencing. Our results show the first example of an RNA-dependent gene _trans_-silencing mediated by epigenetic marks in S. cerevisiae.
[Keywords: RNA, silencing, transcription, chromatin, Set1, Xrn1]
Recent data have shown that cryptic unstable transcripts (CUTs) are RNA polymerase II (RNAPII)-dependent noncoding RNAs (ncRNAs) corresponding to inter- and intragenic regions of the genome and may represent 10% of intergenic transcripts in Saccharomyces cerevisiae (Wyers et al. 2005). Under normal conditions, CUTs are almost undetectable, as they are rapidly degraded by the activity of Rrp6 and Trf4, members of the nuclear exosome and the TRAMP complex, respectively (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005). In addition to Trf4 and Rrp6, the cytoplasmic 5′–3′ exoribonuclease Xrn1 also plays an important role in the turnover of CUTs, supporting the idea that some of these transcripts escape the nuclear quality control and might have a cytoplasmic residency (Thompson and Parker 2007).
Despite these observations, the function(s) of CUTs remain(s) poorly characterized. Cryptic transcription has been widely described from yeast to human and qualified as “transcriptional noise.” Interestingly, it has been proposed that cryptic transcription allows RNA polymerase-dependent chromatin changes but not the production of functional RNA molecules, as those are immediately degraded (Struhl 2007). In agreement with this model, CUT transcription has been shown to interfere with promoters of coding regions and hence regulates gene expression in S. cerevisiae (Martens et al. 2005; Hongay et al. 2006; Uhler et al. 2007). However, alternative models emerged, providing a direct function for cryptic transcripts. Indeed, recent reports have shown that the processing of siRNAs in Schizosaccharomyces pombe is mediated by homologs of the TRAMP and exosome subunits (Buhler et al. 2007; Nicolas et al. 2007), strongly indicating that the fission yeast’s siRNAs might originate from CUTs. It is tempting to hypothesize that CUTs in S. cerevisiae are linked to an ancient mechanism that would control gene expression in an RNA-dependent manner.
One of the RNAi functions is to limit mobile genetic element (MGE) proliferation in the genome (Weiner 2002). MGEs encompass viruses, transposons, and retrotransposons and are dispersed from bacteria to mammalian cells (Capy 2005). The paroxysm of mobile elements has been described in human cells whereby up to 40% of the genome is composed of repeated sequences (Lander et al. 2001) that play potentially deleterious functions in chromosome rearrangements and gene expression. Therefore, each step of the transposition cycle is tightly controlled. In S. cerevisiae, the Ty1 retrotransposon is subjected to a cosuppression regulation (Jiang 2002) that presents similarities with the transcriptional cosuppression mediated by siRNAs in plants (Jorgensen 1995). This observation strongly suggests that a yet-uncharacterized RNAi-like mechanism might exist in budding yeast, despite the absence of the entire RNAi pathway in this organism (Aravind et al. 2000).
In this study, we address the existence of an RNA-dependent gene regulation mechanism in S. cerevisiae. By analogy with transposon regulation in higher eukaryotes, we hypothesized that regulatory RNAs might provide the silencing signal to repress Ty1 expression. With the use of strains defective for the 5′–3′ and 3′–5′ RNA decay pathways, we show the existence of a TY1 unstable antisense RNA that triggers Ty1 transcriptional silencing in trans. We present evidence that the Ty1 silencing is partially suppressed by in vivo treatment with the histone deacetylase inhibitors nicotinamide (NAM) and Trichostatin A (TSA). In addition, the Ty1 silencing is controlled by the histone methyltransferase (HMT) Set1 and the histone H3 Lys 4 methylation, providing an interesting parallel with heterochromatic gene silencing. We propose that CUTs contribute to establish a specialized chromatin domain over repetitive elements in S. cerevisiae to limit their dispersion.
Results
The 5′–3′ exoribonuclease Xrn1 maintains Ty1 expression and mobility and destabilizes a Ty1 antisense RNA
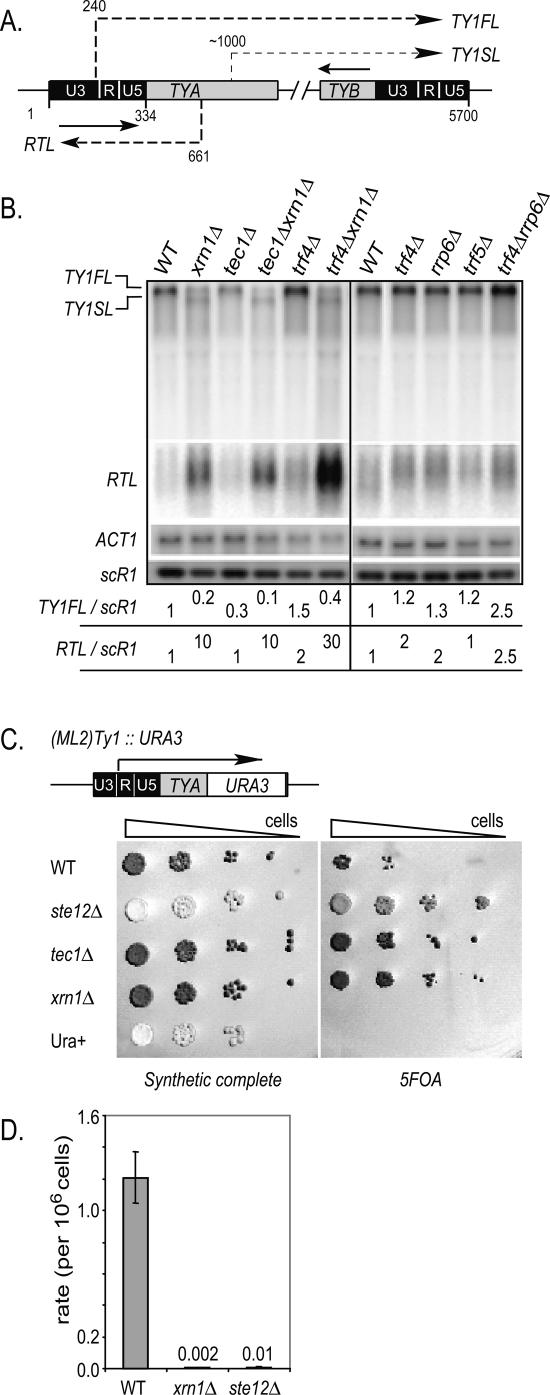
To address the question whether regulatory RNAs can be detected within the Ty1 transposon, we analyzed, by Northern blot, TY1 RNAs in strains mutated for 5′ and 3′ RNA decays that accumulate high levels of CUTs. Thus, total RNAs from yeast strains deleted for XRN1, RRP6, TRF4, or TRF5 encoding genes were extracted, loaded on 1% agarose gel, and transferred onto a nitrocellulose membrane. A _tec1_Δ strain, deleted for a Ty1 transcription activator, was used as a control characterized by negligible levels of TY1 RNA (Laloux et al. 1994; Baur et al. 1997; Morillon et al. 2000). We probed the membrane with sense and antisense riboprobes corresponding to different regions of Ty1 (schematized in Fig. 1A) and normalized the signals with the scR1 RNA level. In the _xrn1_Δ strain, quantifications revealed a dramatic 10-fold accumulation of a >400-nucleotide (nt)-long antisense RNA, mapping to the 5′-end long terminal repeat (LTR) region, and that we named RTL (Fig. 1B). In contrast, _rrp6_Δ and _trf4_Δ showed a modest twofold increase of the RTL RNA, with a slight additive effect in the _rrp6_Δ_trf4_Δ strain (2.5-fold increase). It is interesting to note that the _tec1_Δ_xrn1_Δ strain has similar levels of the RTL RNA as the _xrn1_Δ strain, suggesting that RTL transcription is Tec1-independent. Remarkably, the TY1 full-length sense RNA (TY1FL) showed a fivefold decrease in the _xrn1_Δ strain, similarly to the _tec1_Δ strain (Fig. 1B). In addition, we noticed the presence of a sense-orientated short-length Ty1 RNA (TY1SL) in the _xrn1_Δ strain. Further characterization with a probe spanning the 5′ end of the TYA region failed to reveal the TY1SL RNA, suggesting that TY1SL is a 5′-end-truncated form of the TY1FL RNA (data not shown). Interestingly, the _tec1Δ xrn1_Δ double mutant showed an additive decrease of the TY1FL RNA, suggesting that Xrn1 and Tec1 regulate TY1FL through two independent pathways. In contrast, TRF4, TRF5, and RRP6 deletions only resulted in slight increases of the TY1FL steady-state RNA levels. Taken together, these results show that the cytoplasmic 5′-exonuclease Xrn1 up-regulates the TY1FL RNA but strongly represses the accumulation of two unstable TY1 RNAs: RTL and TY1SL.
Figure 1.
The 5′–3′ exoribonuclease Xrn1 prevents Ty1 antisense RNA accumulation and maintains Ty1 expression and transposition. (A) Schematization of different Ty1 transcripts (dashed lines/arrows), with their respective coordinates. Positions of riboprobes specific to the RTL, TY1FL, and TY1SL RNAs are represented by solid line/arrows. Note that the probe in the TYB region hybridizes the TY1SL and TY1FL RNAs. (B) Northern analysis of Ty1 full-length (TY1FL), Ty1 5′ short-length (TY1SL), and 5′-LTR antisense TY1 RNA (RTL). scR1 and ACT1 were used as loading controls. Numbers represent TY1FL/scR1 and RTL/scR1 RNA ratios. Strains are YAM92 (WT W303), YAM591 (_tec1_Δ), YAM460 (_tec1_Δ_xrn1_Δ), YAM97 (_xrn1_Δ), YAM456 (_trf4_Δ), and YAM458 (_xrn1_Δ_trf4_Δ). YAM118 (WT BY4741), YAM125 (_trf4_Δ), YAM123 (_rrp6_Δ), YAM126 (_trf5_Δ), and YAM128 (_rrp6_Δ_trf4_Δ). (C) Growth assay of strains bearing TY1(ML2)∷URA3 on plates containing complete synthetic media and +5FOA. The strains are YAM698 (WT), YAM700 (_xrn1_Δ), YAM783 (_tec1_Δ), YAM784 (_ste12_Δ), and YAM166 (ura+). (D) Transposition rate of _TY1HIS3AI_-containing strains, expressed as a rate of HIS+ cells per total number of cells (see Materials and Methods). Strains are YAM359 (WT), YAM641 (_xrn1_Δ), and YAM601 (_ste12_Δ). The transposition rate represents the mean of three independent experiments with standard deviations.
To confirm the importance of Xrn1 in Ty1 expression, we constructed a strain carrying the URA3 gene in frame with the highly expressed Ty1(ML2) on chromosome XIII (Morillon et al. 2002). The expression of TY1(ML2)∷URA3 was determined by assessing the resistance of cells to the 5FOA drug (see Materials and Methods). We performed a growth assay dispensing serial 10× cell dilutions of wild-type, _ste12_Δ, _tec1_Δ, and _xrn1_Δ strains carrying Ty1(ML2)∷URA3, on 5FOA plates (Fig. 1C). As expected, the _Ty1(ML2)∷URA3 xrn1_Δ strain was more resistant than the wild type to the 5FOA drug, similar to what is observed in strains mutated for the Ty1 transcription activators Tec1 and Ste12 (Laloux et al. 1994; Baur et al. 1997; Morillon et al. 2000). This result shows that Xrn1 is required for the expression of a single and highly transcribed Ty1 element.
Next, we aimed to test whether Xrn1 influences Ty1 mobility with the use of the Ty1HIS3(AI) strain carrying the HIS3(AI) gene inserted into a chromosomal Ty1 retrotransposon (see Materials and Methods). We observed that the transposition rate of Ty1_HIS3(AI)_ was reduced >500-fold in the absence of Xrn1 compared with the wild type, similar to the rate in the _ste12_Δ strain (Fig. 1D), suggesting that Xrn1 is required for Ty1 mobility.
Altogether these results show that the 5′–3′ exoribonuclease Xrn1 maintains high levels of Ty1 expression and transposition. In addition, Xrn1 controls shorter length TY1 RNAs species, in particular the Ty1 antisense RTL RNA.
RTL RNA is a CUT initiating within the Ty1 coding region
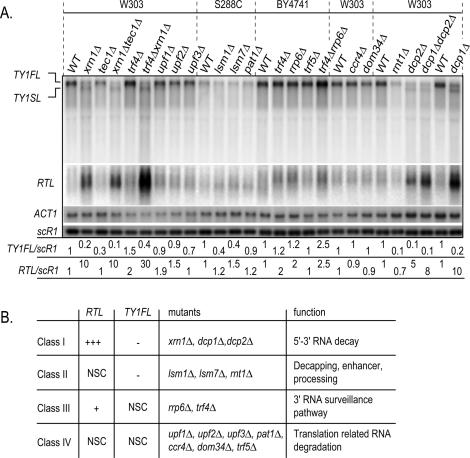
To further characterize the RTL RNA, we extended our analysis to a wider variety of RNA decay mutants. In addition to the _xrn1_Δ mutant, deletions of DCP1 and DCP2 genes encoding the decapping activity (Coller and Parker 2004) showed a 10-fold and fivefold increase of the RTL RNA, respectively (Fig. 2A). In contrast, RTL RNA levels were modestly increased in _pat1_Δ, _lsm1_Δ, and _lsm7_Δ strains, defective for the RNA decapping enhancers (Coller and Parker 2004). Similarly, mutants defective for the No-Go-Decay (_dom34_Δ) (Doma and Parker 2006), for the nonsense-mediated decay (_upf1_Δ, _upf2_Δ, _upf3_Δ) (Muhlrad and Parker 1994), for the RNA-deadenylation pathway (_ccr4_Δ) (Daugeron et al. 2001), and for the endoribonuclease III RNT1 (_rnt1_Δ) (Danin-Kreiselman et al. 2003) showed wild-type levels of the RTL RNA. Interestingly, when comparing the levels of RTL and TY1FL RNAs, all mutants can be divided into four classes (summarized in Fig. 2B). Class I consists of cytoplasmic 5′–3′ RNA decay mutants and is characterized by a large decrease of the TY1FL RNA (more than twofold) concomitant with the appearance of high levels of RTL (more than fivefold). Class II is represented by mutants defective for decapping enhancing activities and nuclear RNA processing and that show a significant reduction of TY1FL (twofold) but without appearance of the RTL RNA (less than twofold). In contrast, class III mutants, defective in the 3′ nuclear surveillance pathway, have no changes in TY1FL but accumulate intermediate levels of RTL (up to fivefold). Finally, class IV is composed of mutants altered in translation-coupled RNA degradation that have very little effect on any Ty1 RNAs.
Figure 2.
Effect of different RNA degradation pathways on Ty1 RNAs. (A) Extended Northern analysis of transcript levels in strains bearing mutations in different RNA surveillance pathways. The membrane is identical as in Figure 1B, but mutants have been separated for presentation convenience. The probes are the same as in Figure 1B. In the left panel, the W303 strains are YAM92 (WT W303), YAM591 (_tec1_Δ), YAM97 (_xrn1_Δ), YAM460 (_tec1_Δ_xrn1_Δ), YAM456 (_trf4_Δ), YAM458 (_xrn1_Δ_trf4_Δ), YAM93 (_upf1_Δ), YAM94 (_upf2_Δ), and YAM95 (_upf3_Δ). W303 strains in the right panel are YAM92 (WT W303), YAM584 (_ccr4_Δ), YAM586 (_dom34_Δ), YAM115 (WT W303), (YAM119), YAM141 (_dcp2_Δ), YAM142 (_dcp1_Δ_dcp2_Δ), YAM92 (WT W303), and YAM96 (_dcp1_Δ). The S288C strains are YAM31 (WT S288C), YAM34 (_lsm1_Δ), YAM33 (_lsm7_Δ), and YAM32 (_pat1_Δ). The BY4741 background strains are YAM118 (WT BY4741), YAM125 (_trf4_Δ), YAM123 (_rrp6_Δ), YAM126 (_trf5_Δ), and YAM128 (_rrp6_Δ_trf4_Δ). (B) Table summarizing the effects of the four classes of mutants on Ty1 RNA accumulation. RTL and TY1FL RNA changes were classified by the following rule: (+++) more than fivefold increase; (+) between twofold and fivefold increase; (−) more than twofold decrease; (NSC) no significant change (less than twofold).
According to this classification, we conclude that the cytoplasmic 5′–3′ RNA decay is the main pathway controlling the RTL RNA stability but independently of translational-related and RNA deadenylation-coupled RNA decay pathways. Interestingly, the 3′ RNA nuclear surveillance machinery plays a minor role in RTL stability except when the 5′–3′ RNA decay is strongly impaired. Finally, we noted that RTL accumulation systematically correlates with the decrease of the steady-state levels of TY1FL RNA, suggesting a causal link for the fate of these two transcripts.
Ty1 RTL RNA initiates within Ty1, is synthesized by RNAPII, and degraded by Xrn1
To define more precisely the RTL RNA, we performed Rapid Amplification of CDNA Ends (5′-RACE) followed by sequencing analysis. It defined several transcription start sites in the TYA coding region with a major starting point at the +661 position from the beginning of the LTR sequence (schematized in Fig. 1A). No open reading frame was identified, suggesting that the TY1 RTL RNA is noncoding.
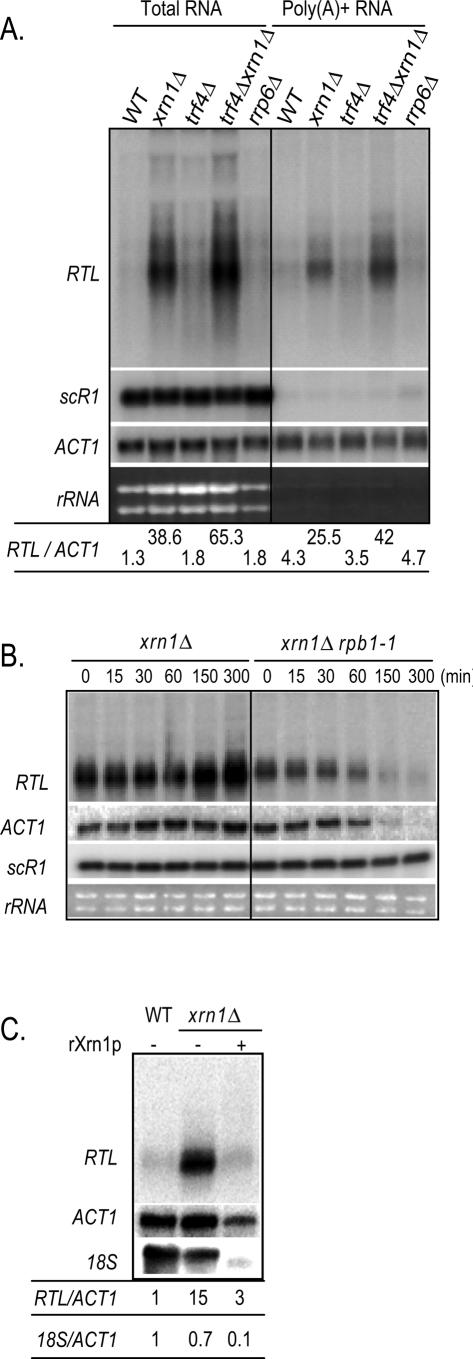
Furthermore, Northern blot analysis of polyadenylated [poly(A)+] RNAs showed that the poly(A)+ RTL RNA is still detectable in the _xrn1_Δ strain (Fig. 3A). Moreover, RTL/ACT1 ratios from total RNAs and poly(A)+ RNAs extracted from the _xrn1_Δ strain suggest that the majority of the RTL RNA is polyadenylated (66%) (Fig. 3A). Unexpectedly, we noted equivalent amounts of poly(A)+ RTL extracted from the _trf4Δ xrn1_Δ mutant (64%). This observation suggests that the additive accumulation of the RTL RNA in the _trf4Δ xrn1_Δ mutant is not related to the Trf4-dependent poly(A) polymerization activity, consistent with previous observations showing that Trf4 has additional roles in RNA stability (Wyers et al. 2005).
Figure 3.
Characterization of the Ty1 antisense RNA. (A) Northern blot analysis of transcript levels in strains bearing mutations in the RNA surveillance pathways, purified on oligo(dT) beads [poly(A+) RNA, right panel] or not (total RNA, left panel). The probes are the same as in Figure 1B. The strains are YAM92 (WT), YAM97 (_xrn1_Δ), YAM456 (_trf4_Δ), YAM458 (_xrn1_Δ_trf4_Δ), and YAM587 (_rrp6_Δ). (B) Northern blot analysis of total RNA upon RNAPII inactivation. The time is in minutes after a 37°C heat shock. The probes are the same as in Figure 1B. The strains are YAM9 (_xrn1_Δ) and YAM538 (xrn1Δrpb1-1). (C) Northern blot analysis of 1 μg of total RNA treated (+) or not (−) with 0.3 μg of recombinant Xrn1p (rXrn1p). The probes are the same as in Figure 1B. The strains are YAM92 (WT) and YAM97 (_xrn1_Δ).
Next, to confirm that the RTL RNA is produced by RNAPII, deletion of XRN1 was performed in a thermosensitive rpb1ts mutant strain (rpb1-1), mutated for RPO21, encoding the main subunit of RNAPII (see Materials and Methods). As shown in Figure 3B, after 60 min at 37°C, RTL was no longer visible in the xrn1Δrpb1-1 strain (see quantification in Supplemental Fig. 1), similarly to the RNAPII-dependent RNA ACT1. In contrast, levels of the RNAPI-dependent rRNA and the RNAPIII-dependent scR1 RNA remained unchanged, suggesting that RNAPII synthesizes the RTL RNA. In addition, the relatively rapid decrease of RTL levels in the xrn1Δrpb1-1 strain suggests that another pathway is involved in RTL degradation, consistent with an additive role of the 3′ nuclear surveillance pathway in RTL stability.
Finally, we addressed the question whether Xrn1 directly degrades the RTL RNA or controls an activator of RTL transcription. It has been shown that Xrn1 preferentially degrades RNA with 5′ monophosphate extremity but not RNA with a capped extremity in vivo and in vitro (Muhlrad et al. 1995). Thus, if RTL is subjected to a Xrn1-dependent degradation in vivo, it should present an uncapped extremity when accumulating in the _xrn1_Δ strain. To test this hypothesis, we treated total RNAs extracted from the _xrn1_Δ strain with purified recombinant yeast rXrn1p and analyzed the degradation profile by Northern blot (Fig. 3C). We observed that the RTL RNA was degraded by rXrn1p with the same efficiency as the uncapped 18S ribosomal RNA (rRNA) (fivefold and sevenfold reduction, respectively). In contrast, the capped ACT1 mRNA was resistant to rXrn1p activity. This result indicates that the RTL RNA detected in the _xrn1_Δ strain is mainly uncapped, confirming that the high accumulation of the RTL RNA is a consequence of RNA stabilization and not transcriptional activation in this strain.
Taken together, our data indicate that RTL is synthesized by RNAPII initiating within the Ty1 coding region and polyadenylated. The Ty1 RTL RNA is then subjected to decapping followed by 5′–3′ RNA degradation performed by Xrn1. RTL destabilization is reinforced by the nuclear Trf4-dependent RNA surveillance pathway.
TY1 RTL RNA controls TY1 expression in trans
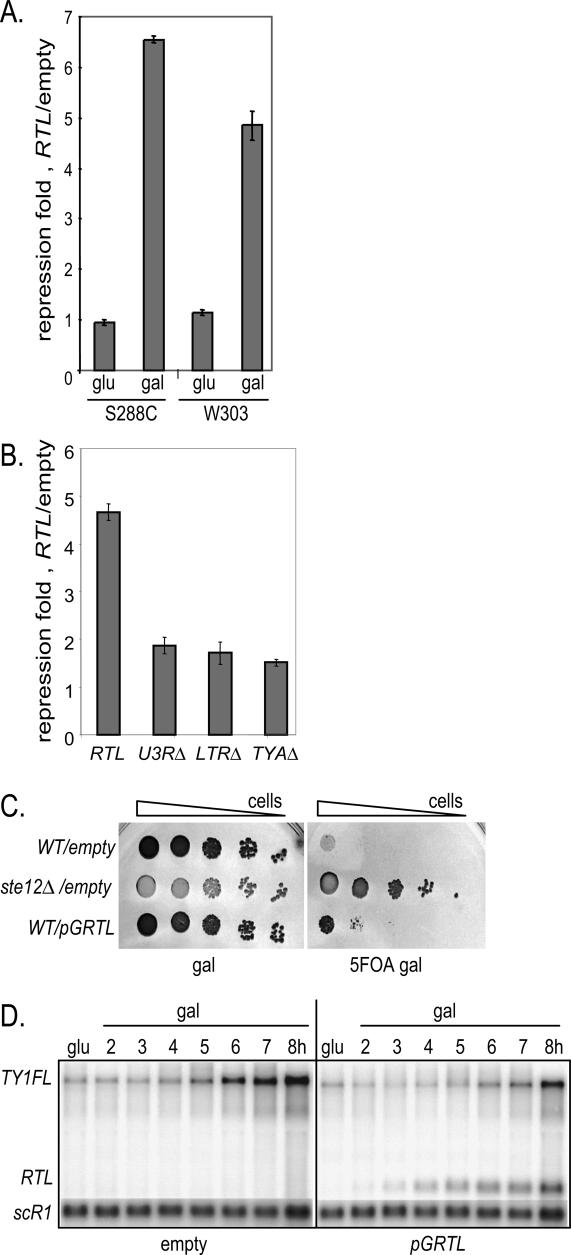
As RTL accumulation coincides with down-regulation of Ty1 expression, we next asked whether the RTL RNA directly controls Ty1 expression and mobility. We cloned the RTL sequence under the _GAL1_-inducible promoter (pGRTL). By switching the carbon source from glucose to galactose in the growth media, we assessed the effect of RTL overexpression on Ty1 mobility. As shown in Figure 4A, the transposition rate of the Ty1HIS3(AI) element was reduced from fourfold to sevenfold in two different yeast genetic backgrounds, W303 and S288C, respectively, upon transcriptional activation of RTL when compared with an empty plasmid. In contrast, upon repressive conditions in glucose, the Ty1HIS3(AI) transposition rate was identical in the strain containing the pGRTL and the strain containing the empty plasmid. We conclude that the RTL RNA inhibits Ty1 retrotransposition in trans.
Figure 4.
_Trans_-silencing of Ty1 expression in the presence of the Ty1 RTL RNA. (A) Transposition repression fold of TY1HIS3AI strains in the presence of pGRTL. The repression fold is expressed as a ratio between the transposition rate measured in a strain bearing pGRTL (pAM134) and a strain bearing an empty plasmid (pAM80) under the same growth conditions. The strains are YAM359 (S288C) and YAM358 (W303), grown in glucose (glu) or galactose (gal). The repression fold represents the mean of three independent experiments with standard deviations. (B) Transposition repression fold of TY1HIS3AI strains in the presence of truncated versions of RTL. The strains are the same as in A but with YAM359 (WT) grown in galactose and bearing pGRTL (RTL), pGRTLU3RΔ (U3RΔ), pGRTLLTRΔ (LTRΔ), or pGRTLTYAΔ (TYAΔ). (C) Growth ability of TY1(ML2)∷URA3 strains containing pGRTL (pAM167) or an empty plasmid (pAM20). Galactose plates contain complete synthetic media without leucine supplemented with 5FOA (5FOAgal) or not (gal). Cells were dropped after serial 10× dilutions. The strains are YAM698 (WT) and YAM784 (_ste12_Δ). (D) Northern blot analysis of total RNAs in a PGAL1ste12 strain (YAM799) bearing pGRTL (pAM134) or an empty plasmid (pAM80). The probes are the same as in Figure 1B. The time is in hours of growth in galactose.
To get insights into the minimal regulatory region(s) of RTL, we cloned, under the control of GAL1 promoter, RTL sequences lacking the U3R (pGRTLU3RΔ), the LTR (pGRTLLTRΔ), and the TYA (pGRTLTYAΔ) (see Supplemental Fig. 2 for a schematic view of the truncated RTL versions). Interestingly, expression of RTLU3RΔ, RTLLTRΔ, or RTLTYAΔ failed to fully repress (less than twofold) Ty1 mobility (Fig. 4B), showing that the integrity of the RTL RNA is required to silence Ty1 transposition. It is interesting to note that the minimal U3R region, corresponding to the Ty1 promoter and 5′ untranslated region of the Ty1 RNA, is required for the Ty1 silencing, suggesting that the RTL RNA acts in trans to control transcription.
Furthermore, to test whether RTL represses a single genomic and highly expressed Ty1 element, we performed a growth test assay on 5FOA plates with the Ty1(ML2)∷URA3 strain transformed with the pGRTL or an empty plasmid (Fig. 4C). We observed that Ty1(ML2)∷URA3 strain grew better on 5FOA+ galactose plates in the presence of pGRTL when compared with the TY1(ML2)∷URA3 strain containing an empty plasmid, but with a lower efficiency than a _Ty1(ML2)∷URA3 ste12_Δ strain, fully 5FOA-resistant. This result confirms that RTL down-regulates the highly expressed Ty1(ML2) in trans by controlling Ty1 expression at transcriptional or/and post-transcriptional steps.
Finally, to kinetically monitor _RTL_-dependent repression on the newly synthesized TY1 RNA, we designed a PGAL1ste12 strain containing the STE12 gene, encoding the Ty1 transcription activator, under the control of the GAL1 promoter. The PGAL1ste12 strain was transformed with the pGRTL plasmid or an empty vector. In glucose-containing medium, repressing STE12 transcription, we observed very low levels of the TY1FL RNA, confirming that STE12 is required for Ty1 transcription (Fig. 4D, first lane in left panel). As expected, upon shift to galactose-containing media and in the presence of the empty vector, we observed progressive increase of the TY1FL RNA levels (Fig. 4D, left panel; see quantification in Supplemental Fig. 3). In contrast, coexpression of RTL impaired the Ste12-dependent TY1 activation, suggesting that the RTL RNA represses the accumulation of neo-synthesized TY1 RNA (Fig. 4D, right panel; see quantification in Supplemental Fig. 3).
Altogether, these results strongly suggest that the RTL RNA silences the Ty1 expression in trans through an RNA-dependent mechanism acting on the de novo synthesized TY1 RNA.
Xrn1 controls Ty1 expression at a post-TBP recruitment step
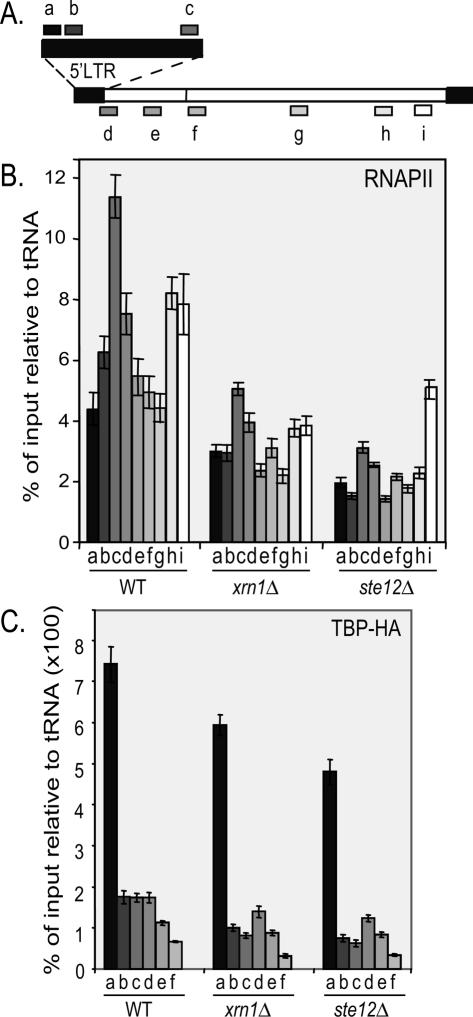
In an attempt to define the RNA-dependent regulatory mechanism that controls Ty1 expression, we performed chromatin immunoprecipitation (ChIP) on formaldehyde-cross-linked cells and recovered the coimmunoprecipitated DNA. The DNA was subjected to real-time PCR by using Ty1-specific probes (schematized on Fig. 5A) to measure changes in the relative occupancy of chromatin-associated factors. When immunoprecipitating Rpb1, the largest subunit of RNAPII, we found that RNAPII occupancy decreased from twofold to threefold within the Ty1 elements in _xrn1_Δ and _ste12_Δ strains when compared with the wild type (Fig. 5B). In contrast, the unrelated constitutive POL1 gene showed equivalent levels of RNAPII occupancy in wild-type, _xrn1_Δ, and _ste12_Δ strains (Supplemental Fig. 4), strongly suggesting that, in the _xrn1_Δ strain, Ty1 transcription is severely impaired and reaches background levels observed in the _ste12_Δ strain.
Figure 5.
Xrn1 controls Ty1 transcription initiation at a post-TBP recruitment step. (A) Positions of different Ty1 amplicons on the Ty1 chromatin for real-time PCR analysis. (B) Real-time PCR quantification of RNAPII using primers over Ty1 chromatin. Percentage of immunoprecipitation signals relative to input, normalized with the tRNA region. The strains are YAM198 (WT), YAM775 (_xrn1_Δ), and YAM778 (_ste12_Δ). (C) Real-time PCR quantification of HA-TBP using primers over Ty1 chromatin. The same normalization as in B. The strains are YAM208 (WT), YAM238 (_xrn1_Δ), and YAM756 (_ste12_Δ).
The transcription process is controlled at the level of initiation, elongation, and termination (Svejstrup 2004). To discriminate which transcription step is controlled upon Ty1 silencing, we performed ChIP experiments using wild-type and _xrn1_Δ strains carrying the HA-tagged TBP, which is associated with the preinitiation complex for RNAPII-dependent regulation (Kuras and Struhl 1999). Remarkably, TBP occupancy was not significantly affected in _xrn1_Δ and _ste12_Δ strains (Fig. 5C), suggesting that TBP recruitment on the Ty1 promoter is not a limiting step when Ty1 is silenced.
Altogether, our results suggest that the RTL RNA promotes Ty1 gene silencing through a transcriptional mechanism occurring after TBP recruitment.
Histone deacetylation and HMT Set1 mediate Ty1 gene silencing
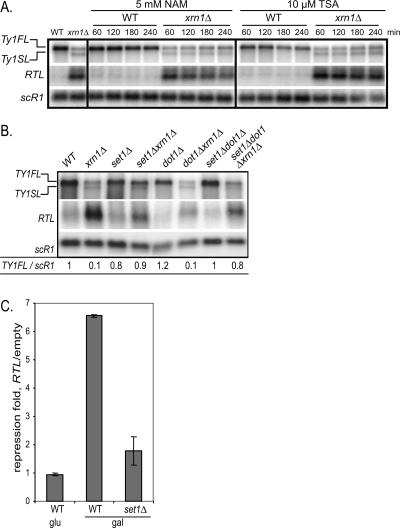
Gene silencing is a key feature of heterochromatin domains and is marked by epigenetic signals such as histone H3 and H4 lysine deacetylation. In addition, HMT Dot1 and Set1 controlling H3K79me and H3K4me, respectively, create a boundary between euchromatin and heterochromatin through a mechanism that might restrict localization of the silencing regulators Sir3 and/or Sir2 (van Leeuwen et al. 2002; Santos-Rosa et al. 2004). To test whether the Ty1 silencing mechanism involves heterochromatin regulators, we first treated wild-type and _xrn1_Δ strains with histone deacetylase inhibitors NAM and TSA, repressing nicotinamide adenine dinucleotide (NAD)-dependent and class I or II histone deacetylases, respectively (Bernstein et al. 2000; Bitterman et al. 2002). Cells treated with 5 mM NAM or 10 μM TSA were harvested at different time points to extract total RNAs. Northern blot analysis (Fig. 6A) showed that, in the _xrn1_Δ strain, the TY1FL RNA levels increased up to threefold after 240 min of NAM treatment (see quantification in Supplemental Fig. 5A). In addition, TSA treatment showed a dramatic eightfold increase of the TY1FL levels in the _xrn1_Δ strain after 240 min (see quantification in Supplemental Fig. 5B). Furthermore, we noted that the RTL RNA levels were twofold decreased upon NAM treatment (see quantification in Supplemental Fig. 5C), suggesting that a NAD-dependent histone deacetylase(s) mediate(s) the Ty1 silencing, putatively by controlling the RTL accumulation. In contrast, upon TSA treatment, the RTL levels increased up to twofold (see quantification in Supplemental Fig. 5C), strongly supporting that a class I or a class II histone deacetylase controls the Ty1 silencing at a step following RTL expression. This analysis suggests that deacetylation of histones is a key regulation step of Ty1 silencing.
Figure 6.
Histone deacetylation and HMT Set1 mediate Ty1 silencing. (A) Northern blot analysis of total RNAs in the presence of histone deacetylase inhibitors. The probes are the same as in Figure 1B. The strains YAM250 (WT) and YAM97 (_xrn1_Δ) were treated with 5 mM NAM or 10 μM TSA, and cells were collected at the indicated time points. (B) Northern analysis of total RNAs. The probes are the same as in Figure 1B. The strains are YAM250 (WT), YAM97 (_xrn1_Δ), YAM249 (_set1_Δ), YAM448 (_set1_Δ_xrn1_Δ), YAM248 (_dot1_Δ), YAM449 (_dot1_Δ_xrn1_Δ), YAM247 (_set1_Δ_dot1_Δ), and YAM444 (_set1_Δ_dot1_Δ_xrn1_Δ). (C) Repression fold of transposition rates of TY1HIS3AI strains bearing the pGRTL and empty plasmids. The strains are YAM359 (WT) and YAM702 (_set1_Δ). The same conditions as in Figure 4A.
Next, to further determine whether Ty1 silencing resembles heterochromatin silencing, _set1_Δ and _dot1_Δ mutants were combined with XRN1 deletion, and total RNAs were probed for TY1FL and RTL RNAs (Fig. 6B). We observed that, in _dot1_Δ_xrn1_Δ, the TY1FL remained at the same level as in the _xrn1_Δ strain (Fig. 6B). In contrast, a null allele of SET1 inserted in the _xrn1_Δ strain restored wild-type TY1FL levels, strongly suggesting that Set1 is required to silence Ty1. However, as with the NAM treatment, we observed a slight decrease of RTL in _set1Δ xrn1_Δ compared with the _xrn1_Δ strain, suggesting that the TY1FL derepression might be due to the decrease of the RTL RNA levels.
To further test the role of Set1 in Ty1 silencing, we asked whether Set1 controls the _RTL_-dependent silencing in trans. The SET1 gene was deleted in the strain containing the Ty1HIS3(AI) to measure the Ty1 mobility in the presence of pGRTL upon galactose induction. As shown in Figure 6C, the _set1_Δ strain containing the pGRTL showed a modest twofold repression of the Ty1 transposition rate when compared with the sevenfold repression in wild type. As the _set1_Δ strain showed mild change in the pGRTL induction (data not shown), we conclude that RTL mainly represses the Ty1 expression via Set1.
Taken together, these results show that Ty1 silencing is mediated by epigenetic signals established by Set1 and histone deacetylases and presents strong similarities with heterochromatin silencing.
Ty1 silencing is controlled by H3K4me
To get further insights into the Ty1 silencing mechanism, we investigated the role of Set1 activity on Ty1 expression. The HMT Set1 catalyzes the addition of one, two, or three methyl groups on the H3K4 (H3K4me1, H3K4me2, and H3K4me3, respectively) and plays multiple roles in DNA repair, transcription, and heterochromatin regulation (Dehe and Geli 2006). In addition, H2B ubiquitylation on Lys 123 (H2BK123-Ub) is required for H3K4me2 and H3K4me3, but not for tethering Set1 on chromatin (Sun and Allis 2002; Dehe et al. 2005; Shahbazian et al. 2005).
As Set1 is required for Ty1 silencing, we first tested, by ChIP experiments, whether H3K4me1, H3K4me2, or H3K4me3 would change on Ty1 chromatin upon Ty1 silencing. Interestingly, the methylation states of H3K4 showed no difference in the _xrn1_Δ and wild-type strains (data not shown), suggesting that H3K4 methylation marks have an indirect or transient effect on Ty1 silencing. Alternatively, Set1 might control Ty1 silencing in a histone-independent manner.
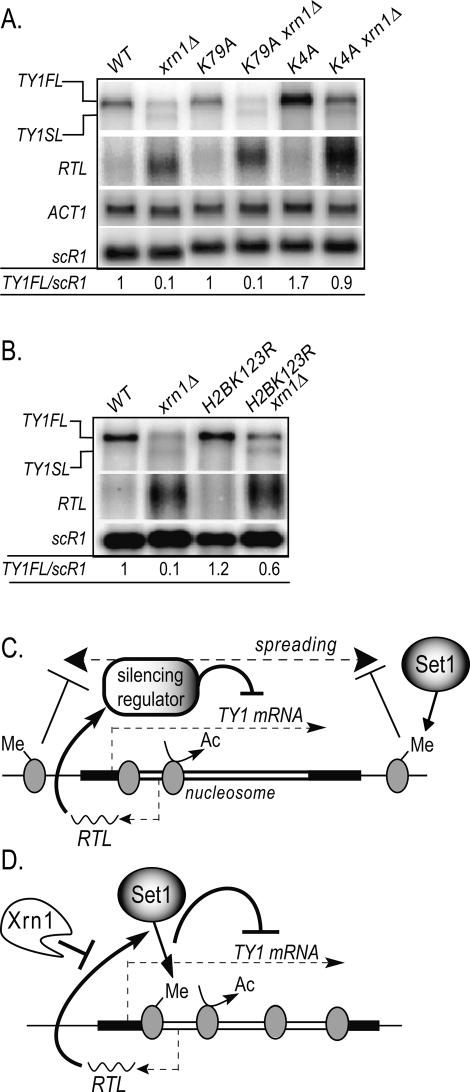
To discriminate between these two possibilities, we asked whether H3K4 methylation is required for the Ty1 silencing. We performed Northern blot analysis with RNAs extracted from strains bearing point mutations at H3 Lys 4. As a control, we used a mutation at H3 Lys 79, targeted by the HMT Dot1. As expected, the _H3K79Axrn1_Δ mutant had similar TY1FL RNA levels as _xrn1_Δ, confirming that Dot1 catalytic activity is not involved in the Ty1 silencing mechanism (Fig. 7A). In contrast, the H3K4A mutant, preventing Set1-dependent H3K4 methylation, showed wild-type levels of TY1FL RNAs when combined with the _xrn1_Δ mutant, supporting the idea that H3K4 methylation promotes Ty1 silencing in the strain lacking Xrn1. In addition, we noted that the H3K4A mutant showed higher levels of the TY1FL RNA than the wild type, in contrast to the _set1_Δ strain (Fig. 6B). This observation suggests that Lys 4 on histone H3 has a residual repressive activity on Ty1 expression even when it is unmethylated.
Figure 7.
H3K4me controls Ty1 silencing. (A) Northern blot analysis of total RNAs. The probes are the same as in Figure 1B. The strains are YAM212 (WT), YAM762 (_xrn1_Δ), YAM216 (H3K4A), YAM764 (H3K4A_xrn1_Δ), YAM214 (H3K79A), and YAM673 (H3K79A_xrn1_Δ). (B) Northern blot analysis of total RNAs. The probes are the same as in Figure 1B. The strains are YAM166 (WT), YAM167 (_xrn1_Δ), YAM835 (H2BK123R), and YAM836 (H2BK123R_xrn1_Δ). (C) Model of RNA-dependent Ty1 transcriptional gene silencing indirectly controlled by Set1. If not destabilized by the cytoplasmic exonuclease Xrn1, the RTL RNA targets an unknown silencing factor that inhibits Ty1 transcription through histone deacetylation. Furthermore, the silencing factor is restricted on Ty1 chromatin by Set1-dependent histone methylation, as suggested for heterochromatic domains. In the absence of Set1, the silencing factor spreads on adjacent chromatin, allowing Ty1 transcription to be derepressed. (D) Model of RNA-dependent Ty1 transcriptional gene silencing directly controlled by Set1. Same as in C but the RTL RNA activates Set1-dependent histone methylation on Ty1 embedded nucleosomes. Histone methylation is then recognized by a putative silencing factor that represses Ty1 transcription through histone deacetylation.
To confirm that Set1 has a histone methylation-dependent activity in Ty1 silencing, we analyzed Ty1 RNA levels in a H2BK123R strain controlling the histone-dependent Set1 activity without impairing its chromatin recruitment. Thus, we combined _xrn1_Δ and H2BK123R mutations and performed a Northern blot analysis (Fig. 7B). Our result showed that the TY1FL RNA level was sixfold increased in _H2BK123Rxrn1_Δ strain compared with _xrn1_Δ, suggesting that H2BK123-dependent histone methylation controls Ty1 silencing. Furthermore, as the H2BK123R strain is only devoid of H3K4me2 and H3K4me3, we propose that Ty1 silencing is mediated by H3K4me2 and/or H3K4me3.
These data support the idea that Set1 controls Ty1 silencing through its histone methylation activity, directly or indirectly.
Discussion
Our results support the existence of an RNA-dependent gene regulation mechanism in S. cerevisiae controlling the Ty1 retrotransposon transcription and mobility. This regulation is mediated by an antisense TY1 RNA, initiating within the TYA region and encompassing the 5′-LTR domain, and necessitates histone deacetylation and Set1-dependent histone methylation, two important epigenetic features of heterochromatin domains. Importantly, the Ty1 silencing is antagonized by the cytoplasmic 5′–3′ exonuclease Xrn1 that prevents accumulation of the cryptic regulatory RNA.
Identification of a novel CUT
In this study, we characterized a novel CUT not identified previously, as repeated sequences were eliminated from the microarray analysis (Wyers et al. 2005). CUTs were shown to accumulate when RNA surveillance pathways are defective, in particular when the Rrp6 component of the exosome is inactive (Wyers et al. 2005). Some CUTs escape the nuclear surveillance pathway but are degraded by the cytoplasmic 5′–3′ decay (Thompson and Parker 2007). This is the case of the Ty1 RTL CUT, identified in this study, which is not subjected to the nuclear surveillance pathway but is very sensitive to the activity of the 5′–3′ decay factors Xrn1, Dcp1 and Dcp2. The nuclear Xrn1-homolog Rat1 has a very modest impact on RTL stability (data not shown), suggesting that RTL processing and degradation are mainly cytoplasmic. The deadenylation coupling RNA degradation factors Ccr4, Pat1, and Lsm proteins have a minor effect on RTL stability, suggesting that RTL might be processed and degraded independently of its poly(A) tail. However, in the absence of the cytoplasmic 5′–3′ RNA degradation pathway, Trf4 plays an important role in the RTL destabilization process as we noted an additive accumulation of the RTL RNA in _xrn1Δ trf4_Δ, implying that these two pathways communicate with each other despite functioning in two different compartments of the cell.
CUTs mediate gene regulation
The function of CUTs remains elusive, but cryptic transcription was proposed to mediate chromatin changes controlling promoter activity and gene expression. In S. cerevisiae, several examples have already been described, and the current model proposes that the CUT transcription interferes with the gene transcription (Martens et al. 2005; Hongay et al. 2006; Uhler et al. 2007). This model implies that the RNA molecule itself has a minor function as being immediately degraded by the nuclear surveillance pathway in normal conditions. Our data show that RTL silences Ty1 expression in trans by acting on the de novo synthesized TY1 RNA. Furthermore, attempts to define a minimalist Ty1 regulatory RNA showed that the TYA and U3/R regions are required for Ty1 silencing, suggesting that the integrity of the regulatory RNA is important. Consistent with this observation, we failed so far to detect shorter, processed regulatory TY1 RNAs that could be involved in the Ty1 silencing in vivo. To our knowledge, this is the first example of RNA-dependent gene _trans_-silencing in S. cerevisiae.
Transcriptional gene silencing
Gene _trans_-silencing has been well characterized in plants and mammalian cells, where it mainly acts at a post-transcriptional level, controlling cognate mRNA translation or degradation. Here, we show that the Ty1 _trans_-silencing affects transcription at early steps. Indeed, two lines of evidence support a transcriptional gene silencing model. First, Ty1 silencing is characterized by a significant reduction of RNAPII occupancy over the Ty1 chromatin, which might lead to a large decrease in Ty1 expression and mobility. Furthermore, we show that histone deacetylation and Set1-dependent histone methylation control Ty1 repression, suggesting that Ty1 silencing is a nuclear event occurring at the chromatin level.
However, we cannot rule out the possibility that Ty1 is silenced additionally at post-transcriptional step(s). This idea is reinforced by the large difference in the transposition repression fold observed in the _xrn1_Δ strain (500-fold) and in a strain in which RTL transcription is induced from a plasmid (sevenfold in presence of pGRTL). This observation suggests that Xrn1 either destabilizes other Ty1 regulatory RNAs or controls post-transcriptional steps during the transposition cycle. For instance, previous studies revealed a structural role of Xrn1 and P-body components in the assembly of gypsy-like Ty3 VLPs (Beliakova-Bethell et al. 2006). One could hypothesize that this function is conserved for the assembly process of Ty1 VLPs as well. This defect would add up to the transcriptional defect we observed in the _xrn1_Δ strain and would provide an explanation for the low transposition rate in this mutant.
The transcriptional silencing model implies that the regulatory TY1 RNA shuttles back to the nucleus. This dynamic model is supported by very few examples in S. cerevisiae. For instance, only some tRNAs have been shown to cycle back to the nucleus after delivering their amino acids under stress conditions (Dahlberg and Lund 2005). Alternatively, we can hypothesize that the Ty1 regulatory RNA is retained and processed in the nucleus, despite Xrn1 and Dcp1/2 proteins being mainly cytoplasmic. In support of this idea, decapping activity was shown to occur in the nucleus when mRNA export was blocked (Kufel et al. 2004). Further experiments on localization of the Ty1 regulatory RNA will provide insights into mechanisms of the Ty1 silencing as well as RNA transport.
Chromatin-mediated gene silencing
Ty1 silencing is mediated by epigenetic signals that control heterochromatin domains. In particular, we showed that the Set1-dependent histone methylation has a crucial role. Set1 has been extensively characterized over the last 5 years (Dehe and Geli 2006). It is part of the eight-subunit SET1C complex that mono-, di-, and trimethylates histone H3 and nonhistone proteins (Roguev et al. 2001; Zhang et al. 2005). H3K4me3 is a mark of active genes, but, surprisingly, no major transcription defect has been linked with the absence of H3K4me3 (Santos-Rosa et al. 2002). Originally, Set1 was characterized to establish subtelomeric HM and rDNA silencing (Nislow et al. 1997). Current models propose that H3K4 methylation restricts silencing factors such as Sir3 to telomeric regions (Santos-Rosa et al. 2004). Indeed, in the absence of Set1, Sir3 spreads into the euchromatin, fading the silencing at the heterochromatin loci. Similarly to heterochromatin silencing, we present evidence that Set1 and H3K4me2 and/or H3K4me3 are also involved in Ty1 silencing. Two alternative models of Ty1 silencing can be proposed.
By analogy with heterochromatin, Set1-dependent H3K4 methylation, flanking the transposon, might restrict a Ty1 putative silencing factor on the transposon. The absence of Set1 and H3K4 methylation would release the silencing factor from the Ty1 chromatin and derepress the Ty1 expression (see model in Fig. 7C). As in heterochromatin domains, histone deacetylases would deacetylate Ty1 chromatin and contribute to Ty1 silencing. In favor of this model for an indirect role of Set1, no change in H3K4 methylation was observed on Ty1 chromatin upon silencing (data not shown).
The second model is based on a direct role of histone H3K4 methylation in Ty1 silencing. The RTL regulatory RNA could transiently target or activate Set1, allowing temporally H3K4 methylation of the Ty1 chromatin that might be subsequently recognized by a specific silencing regulator (see model in Fig. 7D). As for the previous model, histone deacetylation would reinforce the Ty1 silencing. Despite the fact that H3K4me3 and H3K4me2 are marks of actively transcribed chromatin, recent data suggest that they might also have inhibitory effects as well. Indeed, chromatin repressing factors, such as histone deacetylase complexes, were shown to bind directly to the trimethylated H3K4 via the plant homeodomain (PHD) (Mellor 2006; Sims and Reinberg 2006). Moreover, upon silencing induction, H3K4 has been shown to be transiently mono- or dimethylated on the silenced HM regions in S. cerevisiae, revealing a putative direct role of these marks during heterochromatin formation (Katan-Khaykovich and Struhl 2005).
Interestingly, both models involve histone deacetylation. In S. cerevisiae, heterochromatin establishment and maintenance is mainly regulated by the NAD-dependent histone deacetylase Sir2 (Shore et al. 1984), but also by class I and II histone deacetylases (Rundlett et al. 1996). Further investigation will allow us to determine the histone substrate and histone deacetylase(s) involved in Ty1 silencing (Hickman and Rusche 2007).
Ancient form of RNA-dependent gene silencing
Long or short regulatory RNAs have been widely described in prokaryotes and eukaryotes to control gene expression. Following our observations, several similarities between eukaryotic RNA-dependent gene silencing and the _RTL_-dependent Ty1 silencing can be drawn.
Long ncRNAs control gene expression and can initiate chromatin modifications on specific regions. Xist ncRNA and the Hox antisense ncRNAs (HOTAIR RNA) silence the entire X chromosome and the HOX genes’ expression, respectively (Masui and Heard 2006; Rinn et al. 2007). The relationships between the Xist RNA and chromatin modifications are still poorly characterized. But recently, the HOTAIR regulatory RNA was shown to directly interact with the HMT PRC2 to tether and induce histone H3K27 methylation over the Hox locus and hence to control, in trans, gene expression and chromatin organization (Rinn et al. 2007). This direct interaction between a regulatory RNA and a chromatin modifier is fundamental and provides an important step to understand targeted chromatin modifications. Our results show that Set1 is involved in the RNA-dependent Ty1 silencing, and one could speculate that the RTL long ncRNA would directly interact with Set1 protein, targeting its activity to the Ty1 chromatin for silencing.
Concerning short regulating RNAs, we could not provide any data that support a Ty1 silencing mechanism mediated by siRNAs. Nevertheless, it is interesting to note that two genetic screens in Caenorhabditis elegans showed that the Set1 homolog regulates transposon silencing in an RNAi-dependent manner, through a mechanism that could be similar to the Set1-mediated Ty1 silencing described in this study (Grishok et al. 2005; Robert et al. 2005). More strikingly, in rat testis, small regulatory RNAs, such as rasiRNAs and piRNAs, were recently found to control transposable element transcription (O’Donnell and Boeke 2007). Interestingly, these RNAs were proposed to originate from long antisense ncRNA (Vagin et al. 2006) that could resemble the RTL RNA regulating Ty1 expression. Although most of the RNAi features are absent in S. cerevisiae, our work might have uncovered a primitive and ancestral pathway of RNA-dependent gene regulation.
Materials and methods
Yeast strains and plasmid construction
The strains used in this study are described in Supplemental Table 1. The experiments were mostly performed in the W303 background. Gene deletions and epitope tags were introduced by transformation of PCR fragments generated with specific primers (sequences can be obtained upon request) using the appropriate plasmids listed in Supplemental Table 2 (Longtine et al. 1998).
The yeast strain carrying ML2∷URA3 is a derivative of W303 in which we introduced URA3 in frame with TYA(ML2) by single-step recombination. The recombination sites correspond to TYA(967–1007) and to a region located downstream from Ty1(ML2) (chrXIII coordinates 202532–202571) resulting in the deletion of the TYB(ML2) region. Plasmid pAM134 is a derivative of pAM80 (pYESdest52; Invitrogen) in which we introduced the Ty1 sequence amplified by PCR from pAM22 (661–1 nucleotides) following the manufacturer’s instructions. Similarly, partially truncated RTL sequences were cloned to produce pAM182 (_U3R_Δ, 661–290, pGRTLU3RΔ), pAM184 (_TYA_Δ, 334–1, pGRTLTYAΔ), and pAM183 (_LTR_Δ, 661–334, pGRTLLTRΔ). Ty1 sequences were amplified by PCR from pAM3. pAM167 is a derivative of pAM134 with LEU2 instead of URA3. All constructs were subsequently sequenced.
Media and culture conditions
Growth media and plates were prepared with standard methods using YPDA and CSM media (Gibco) supplemented as indicated and containing 2% glucose or 2% galactose. 5FOA experiments were carried out as follows: Transformed yeast cells were grown on appropriate selective media and were dropped on selective CSM plates containing 1 mg/mL 5FOA (Melford laboratory). Histone deacetylase inhibitors were added to YPDA media at 5 mM for NAM (Sigma) and 10 μM for TSA (Sigma).
RNA extraction, poly(A)+ RNA purification, and Northern blotting
Total RNA was extracted using the hot phenol extraction procedure. Poly(A)+ RNAs were purified on oligo(dT) Dynabeads (Invitrogen). Otherwise stated, 5 μg of total or poly(A)+ RNAs were loaded on denaturing 1% agarose gel and transferred to nitrocellulose membranes. Membranes were UV-cross-linked and hybridized overnight at 65°C with either 32P-labeled DNA probes or single-stranded riboprobes in Ultrahyb buffer (Sigma). Blots were washed twice with 0.5× SSC and 1% SDS for 20 min at 65°C. DNA probes were obtained by random-primed labeling (Stratagene) of specific DNA fragments generated by PCR. Single-stranded riboprobes were obtained by SP6 in vitro transcription of gene-specific PCR fragments containing the SP6 promoter (Ambion). PCR primers are available upon request. ACT1 and scR1 RNAs were used as loading controls.
Transposition test
In the Ty1HIS3AI strain, HIS3 was inserted in the TYB region of a Ty1 element in the antisense orientation (Curcio and Garfinkel 1991). HIS3 RNA synthesis was independent of the Ty1 transcription but was interrupted by an artificial intron only spliced during TY1 RNA transcription. The colony became HIS+ when the Ty1_HIS3_(AI) transposed resulting in a newly inserted Ty1_HIS3_ with a functional HIS3 gene. Thus, the transposition rate corresponded to the number of HIS+ over the total number of colonies. Three independent cultures of strains—YAM359 (WT), YAM627 (_xrn1_Δ), and YAM601 (ste12_Δ)—were grown in YPDA overnight at 30°C. Cells were diluted 1:100 in YPDA media and grown 48 h at 20°C. An equal volume of 2× YPDA broth was added, and cultures were grown for an additional 24 h at 20°C. Aliquots of each culture were plated on YPDA and CSM-HIS plates and incubated during 3–4 d at 30°C. The rate of Ty1_his3AI transposition is the number of His+ colonies divided by the total number of colonies plated (on YPDA plates) (Curcio and Garfinkel 1991).
In the _trans_-silencing transposition assay with RTL_-dependent repression, three independent transformants of plasmids pAM80 or pAM134 in strains YAM359 (S288C) and YAM358 (W303) were grown in CSM-Ura 2% glucose broth at 30°C. Cells were diluted 1:100 in CSM-Ura 2% galactose broth and grown for 48 h at 20°C. Glucose was added to reach 2% final, and cells were grown for an additional 24 h at 20°C. Aliquots of each culture were plated on CSM-Ura 2% glucose agar and CSM-Ura-His 2% glucose agar and incubated at 30°C. The frequency of Ty1_his3AI transposition is the number of His+ Ura+ colonies divided by the total number of Ura+ colonies plated.
ChIP
ChIPs were performed essentially as described previously. Yeast strains were grown to OD600 = 0.5 either in YPDA or CSM minimum media at 30°C, and cross-linked for 20 min by the addition of formaldehyde to a final concentration of 1.2%. The cross-linking reaction was quenched by adding glycine at 0.3 M final. Chromatin was sonicated to obtain 400–500 nt of DNA fragment, and 150 μL (at 1.25 μg/μL) of sonicated chromatin was immunoprecipitated overnight in the presence of specific antibodies against the N-terminal domain of Rpb1 (Y80, Santa Cruz Biotechnologies), the HA epitope (fp7, Santa Cruz Biotechnologies) for the TBP HA-tagged strain. Immunoprecipitated chromatin was purified with 5 mg of protein A-Sepharose (Amersham Pharmacia). All immunoprecipitations were repeated at least twice with different chromatin extracts. Immunoprecipitated DNA was quantified by real-time PCR using the LightCycler 480 (Roche), with primer pairs available upon request. Signals are expressed as the percentage of input DNA relative to the tf(GAA)P2 (tRNA region in ChrXVI, coordinates 622628–622537). Error bars correspond to standard deviations.
Acknowledgments
We thank L. Bénard for purified recombinant Xrn1, and A. Johnson, A. Culbertson, G. Chanfreau, D. Libri, B. Séraphin, V. Géli, M.J. Curcio, and D. Garfinkel for kindly providing strains and plasmids. We gratefully thank J. Mellor for constant support and encouragement, allowing this study to initiate, and H. Lehir, D. Libri, J. O’Sullivan, B. Séraphin, F. Stutz, and E. Van Dijk for helpful discussions. Special thanks to Sylvie Carmet for editorial assistance and Lucile Astorgues-Xéri and Violaine St Andrée for technical assistance. Our financial support is from ARC, FRM, ATIP-CNRS, and HFSPO.
Footnotes
References
- Aravind L., Watanabe H., Lipman D.J., Koonin E.V. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl. Acad. Sci. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur M., Esch R.K., Errede B. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol. Cell. Biol. 1997;17:4330–4337. doi: 10.1128/mcb.17.8.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova-Bethell N., Beckham C., Giddings T.H., Winey M., Parker R., Sandmeyer S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 2006;12:94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Tong J.K., Schreiber S.L. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman K.J., Anderson R.M., Cohen H.Y., Latorre-Esteves M., Sinclair D.A. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Buhler M., Haas W., Gygi S.P., Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Capy P. Classification and nomenclature of retrotransposable elements. Cytogenet. Genome Res. 2005;110:457–461. doi: 10.1159/000084978. [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Curcio M.J., Garfinkel D.J. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg J., Lund E. tRNA turnaround. Mol. Cell. 2005;19:292–294. doi: 10.1016/j.molcel.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Danin-Kreiselman M., Lee C.Y., Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- Daugeron M.C., Mauxion F., Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehe P.M., Geli V. The multiple faces of Set1. Biochem. Cell Biol. 2006;84:536–548. doi: 10.1139/o06-081. [DOI] [PubMed] [Google Scholar]
- Dehe P.M., Pamblanco M., Luciano P., Lebrun R., Moinier D., Sendra R., Verreault A., Tordera V., Geli V. Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B. J. Mol. Biol. 2005;353:477–484. doi: 10.1016/j.jmb.2005.08.059. [DOI] [PubMed] [Google Scholar]
- Doma M.K., Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Sinskey J.L., Sharp P.A. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes & Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M.A., Rusche L.N. Substitution as a mechanism for genetic robustness: The duplicated deacetylases Hst1p and Sir2p in Saccharomyces cerevisiae. PLoS Genet. 2007;3:e126. doi: 10.1371/journal.pgen.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay C.F., Grisafi P.L., Galitski T., Fink G.R. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Jiang Y.W. Transcriptional cosuppression of yeast Ty1 retrotransposons. Genes & Dev. 2002;16:467–478. doi: 10.1101/gad.923502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R.A. Cosuppression, flower color patterns, and metastable gene expression states. Science. 1995;268:686–691. doi: 10.1126/science.268.5211.686. [DOI] [PubMed] [Google Scholar]
- Katan-Khaykovich Y., Struhl K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 2005;24:2138–2149. doi: 10.1038/sj.emboj.7600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J., Bousquet-Antonelli C., Beggs J.D., Tollervey D. Nuclear pre-mRNA decapping and 5′ degradation in yeast require the Lsm2-8p complex. Mol. Cell. Biol. 2004;24:9646–9657. doi: 10.1128/MCB.24.21.9646-9657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L., Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Laloux I., Jacobs E., Dubois E. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res. 1994;22:999–1005. doi: 10.1093/nar/22.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Martens J.A., Wu P.Y., Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes & Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui O., Heard E. RNA and protein actors in X-chromosome inactivation. Cold Spring Harb. Symp. Quant. Biol. 2006;71:419–428. doi: 10.1101/sqb.2006.71.058. [DOI] [PubMed] [Google Scholar]
- Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Morillon A., Springer M., Lesage P. Activation of the Kss1 invasive-filamentous growth pathway induces Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:5766–5776. doi: 10.1128/mcb.20.15.5766-5776.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A., Benard L., Springer M., Lesage P. Differential effects of chromatin and Gcn4 on the 50-fold range of expression among individual yeast Ty1 retrotransposons. Mol. Cell. Biol. 2002;22:2078–2088. doi: 10.1128/MCB.22.7.2078-2088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C.J., Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E., Yamada T., Cam H.P., Fitzgerald P.C., Kobayashi R., Grewal S.I. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 2007;14:372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- Nislow C., Ray E., Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K.A., Boeke J.D. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V.J., Sijen T., van Wolfswinkel J., Plasterk R.H. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes & Dev. 2005;19:782–787. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A., Schaft D., Shevchenko A., Pijnappel W.W., Wilm M., Aasland R., Stewart A.F. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen A.A., Kobayashi R., Bavykin S., Turner B.M., Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Bannister A.J., Dehe P.M., Geli V., Kouzarides T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 2004;279:47506–47512. doi: 10.1074/jbc.M407949200. [DOI] [PubMed] [Google Scholar]
- Shahbazian M., Zhang K., Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Shore D., Squire M., Nasmyth K.A. Characterization of two genes required for the position-effect control of yeast mating-type genes. EMBO J. 1984;3:2817–2823. doi: 10.1002/j.1460-2075.1984.tb02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R.J., Reinberg D. Histone H3 Lys 4 methylation: Caught in a bind? Genes & Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Sun Z.W., Allis C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q. The RNA polymerase II transcription cycle: Cycling through chromatin. Biochim. Biophys. Acta. 2004;1677:64–73. doi: 10.1016/j.bbaexp.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Thompson D.M., Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhler J.P., Hertel C., Svejstrup J.Q. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc. Natl. Acad. Sci. 2007;104:8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vanacova S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F., Gafken P.R., Gottschling D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- Weiner A.M. SINEs and LINEs: The art of biting the hand that feeds you. Curr. Opin. Cell Biol. 2002;14:343–350. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- Wyers F., Rougemaille M., Badis G., Rousselle J.C., Dufour M.E., Boulay J., Regnault B., Devaux F., Namane A., Seraphin B., et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Zhang K., Lin W., Latham J.A., Riefler G.M., Schumacher J.M., Chan C., Tatchell K., Hawke D.H., Kobayashi R., Dent S.Y. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]