The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein Modifications: Beyond the Usual Suspects' Review Series (original) (raw)
Abstract
As a lysosomal/vacuolar degradative pathway that is conserved in eukaryotic organisms, autophagy mediates the turnover of long-lived proteins and excess or aberrant organelles. The main characteristic of autophagy is the formation of a double-membrane vesicle, the autophagosome, which envelops part of the cytoplasm and delivers it to the lysosome/vacuole for breakdown and eventual recycling of the degradation products. Among the approximately 30 autophagy-related (Atg) genes identified so far, there are two ubiquitin-like proteins, Atg12 and Atg8. Analogous to ubiquitination, Atg12 is conjugated to Atg5 by Atg7—an E1-like protein—and Atg10—an E2-like protein. Similarly, Atg7 and Atg3 are the respective E1-like and E2-like proteins that mediate the conjugation of Atg8 to phosphatidylethanolamine. Both Atg12–Atg5 and Atg8 localize to the developing autophagosome. The Atg12–Atg5 conjugate facilitates the lipidation of Atg8 and directs its correct subcellular localization. Atg8–phosphatidylethanolamine is probably a scaffold protein that supports membrane expansion and the amount present correlates with the size of autophagosomes.
Keywords: lysosome, membrane biogenesis, protein targeting, stress, vacuole
Glossary
Ape1 aminopeptidase I
Cvt cytoplasm-to-vacuole targeting
GABARAP γ-aminobutyric-acid-type-A-receptor-associated protein
GATE16 Golgi-associated ATPase enhancer of 16 kDa
GFP green fluorescent protein
HECT homologous to the E6AP carboxyl terminus
LC3 microtubule-associated protein 1 light chain 3
PE phosphatidylethanolamine
RING really interesting new gene
Introduction
In eukaryotic organisms, the functional diversity of the protein repertoire can be greatly extended by post-translational modifications. These change the properties of target proteins by modulating their activity, localization, degradation and interaction with other proteins. In this process, the target proteins are covalently modified with other molecules such as ubiquitin. A family of small proteins related to ubiquitin—known as the ubiquitin-like proteins (Ubls)—which can be covalently attached to other proteins has been recently characterized (Kerscher et al, 2006). Among the approximately 10 Ubl family members identified so far are two autophagy-related proteins, Atg8 and Atg12, which are specifically involved in this conserved degradative pathway in eukaryotic organisms. In the past decade, the molecular mechanism of autophagy has begun to be elucidated, leading to the identification of approximately 30 genes that are specifically required for this pathway (Xie & Klionsky, 2007). These are referred to as autophagy-related (Atg) genes. Among them, eight are implicated directly in the Atg8 and Atg12 conjugation systems. Although Atg8 and Atg12 have no clear sequence homology to ubiquitin, their crystal structures reveal a conserved ubiquitin-fold region (Sugawara et al, 2004; Suzuki et al, 2005; Fig 1). Furthermore, these two proteins are attached to their substrate though enzymatic pathways that are similar to the ubiquitin system. Here, we focus on the molecular mechanism of the Atg8 and Atg12 conjugation process and its regulatory network.
Figure 1.
Structural comparisons of ubiquitin, LC3 and _At_Atg12. (A) Ribbon diagrams of ubiquitin (Vijay-Kumar et al, 1987; Protein Data Bank (PDB) code ), LC3 (Sugawara et al, 2004; PDB code ) and _At_Atg12 (Suzuki et al, 2005; PDB code ) are shown in the same orientation; α-helices are shown in purple, 310 helices in blue, β-strands in yellow, β-turns in cyan and unstructured loops in white. (B) Superimposition of ubiquitin (red), LC3 (blue) and _At_Atg12 (green). A multiple structural alignment was constructed using MAMMOTH-mult (http://ub.cbm.uam.es/mammoth/mult; Lupyan et al, 2005) and the structures were visualized with the Visual Molecular Dynamics (VMD) program (http://www.ks.uiuc.edu/Research/vmd). At, Arabidopsis thaliana; Atg, autophagy-related; LC3, microtubule-associated protein 1 light chain 3.
An overview of autophagy
Autophagy mediates the lysosome-dependent turnover of macromolecules and entire organelles. Induced by environmental changes, such as nutrient depletion, cytosolic components are delivered to the vacuole (the lysosome analogue in yeast) through the autophagy pathway and are degraded by resident hydrolases. Degradation products are recycled to allow the synthesis of new macromolecules. Autophagy also has an important role in various cellular processes and is associated with many human diseases (Huang & Klionsky, 2007; Shintani & Klionsky, 2004).
There are several forms of autophagy, including macroautophagy, microautophagy and chaperone-mediated autophagy. In contrast to the other two types of autophagy, macroautophagy involves de novo vesicle formation that takes place separate from the lysosomal/vacuolar membrane. In this process, cytosolic components are sequestered in a double-membrane vesicle known as an autophagosome. The outer membrane of the autophagosome fuses with the lysosome/vacuole allowing access to the inner contents of the vesicle, which are subsequently degraded. As the mechanism of macroautophagy is better characterized than the other types of autophagy, it is hereafter referred to as autophagy for the purposes of this review. Autophagy has been studied in mammalian cells since the 1950s, and in the 1990s this degradative pathway was reported in the yeast Saccharomyces cerevisiae. Since then, mutant screening in fungi has led to the identification of molecular components of autophagy, the Atg genes (Klionsky et al, 2003). Orthologues of the Atg genes have recently been found and functionally characterized in higher eukaryotes, implying a conserved mechanism.
Autophagosome formation can be divided into three steps: nucleation, expansion and completion. The initial sequestering compartment is called the phagophore, which expands, probably through vesicular addition, to envelop portions of the cytoplasm. The expanding phagophore eventually matures into the autophagosome. In S. cerevisiae, the phagophore-assembly site (PAS; also known as the pre-autophagosomal structure) is proposed to be the site from which the autophagosome originates (Kim et al, 2002; Suzuki et al, 2001). Most Atg proteins co-localize at this site, presumably to participate in vesicle formation in a concerted manner. Accordingly, the PAS can be regarded as a hybrid of Atg proteins and developing vesicles. In yeast, the PAS appears to be perivacuolar and, in most cases, there is one PAS per cell. In mammalian cells, the co-localization of ATG proteins has been observed at multiple loci (Mizushima et al, 2003; Mizushima et al, 2001; Young et al, 2006), probably corresponding to multiple vesicle-formation sites.
Primarily identified as a non-selective degradative pathway, autophagy can also mediate specific cargo delivery. The best studied example is a biosynthetic pathway called the cytoplasm-to-vacuole targeting (Cvt) pathway, which transports the resident vacuolar hydrolase Ape1 into the vacuole (Nair & Klionsky, 2005). Autophagy also has a role in the homeostatic control of unwanted organelles such as excess peroxisomes and mitochondria, which are specifically delivered to the vacuole for degradation through pexophagy and mitophagy, respectively (Kim et al, 2007; Sakai et al, 2006).
Atg12 conjugation system
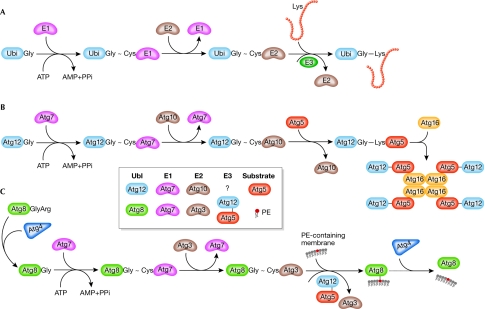
Atg12 was the first ubiquitin-like Atg protein to be identified. In the canonical system, ubiquitin is synthesized as a precursor and is processed by a specific protease to expose the carboxy-terminal glycine residue. Activated by an E1 enzyme, ubiquitin is then transferred to an E2 enzyme, forming a thioester bond. An E3 ubiquitin ligase recognizes the target protein and transfers ubiquitin from the E2 to a lysine residue on the target (Fig 2A).
Figure 2.
Conjugation processes in yeast. (A) Ubiquitin. (B) Atg12. (C) Atg8. See text for details. Atg, autophagy-related; PE, phosphatidylethanolamine; PPi, pyrophosphate; Ubi, ubiquitin; Ubl, ubiquitin-like.
The amino-acid sequence of Atg12 ends with a glycine residue and there is no protease involved in Atg12 conjugation. Analogous to ubiquitination, there is an E1-like enzyme, Atg7, and Atg12 is activated by forming a thioester bond between the C-terminal Gly 186 of Atg12 and the Cys 507 of Atg7 (Tanida et al, 1999). After activation, Atg12 is transferred to Atg10, which is an E2 enzyme (Shintani et al, 1999), and is eventually conjugated to the target protein Atg5 at Lys 149 through an isopeptide bond (Mizushima et al, 1998a; Fig 2B). There is no typical E3 enzyme involved in Atg12–Atg5 conjugation. Atg5 interacts further with a small coiled-coil protein, Atg16, and Atg12–Atg5-Atg16 forms a multimeric complex through the homo-oligomerization of Atg16 (Mizushima et al, 1999). The molecular weight of this multimeric complex is approximately 350 kDa and it probably represents a tetramer of Atg12–Atg5-Atg16 (Kuma et al, 2002).
Although the overall sequence of Atg7 shows little similarity to E1 enzymes in yeast, and so far only Atg7 has been shown to function in the form of a homodimer (Komatsu et al, 2001), Atg7 shares a conserved metal-binding motif and a downstream active-site cysteine residue with other E1 enzymes such as Uba2, Uba3, Uba4 and Uba5. The ATP-binding domain of Atg7 is also homologous to the corresponding region in other E1 enzymes and is essential for the formation of the Atg12–Atg5 conjugate. In contrast to ubiquitin, which is conjugated to multiple targets in an inducible and reversible manner, Atg5 seems to be the only target of Atg12 and the conjugation of Atg12–Atg5 occurs constitutively (Mizushima et al, 1998a). In addition, no processing enzyme has been identified that cleaves the isopeptide bond between Atg12 and Atg5, suggesting that this conjugation is irreversible.
Orthologues of each component of the Atg12 system have been found in mice and humans, and they function similarly to their yeast counterparts (Mizushima et al, 1998b; Mizushima et al, 2002; Tanida et al, 2001). There is also a mammalian Atg16-like protein (ATG16L) that mediates the formation of an ATG12–ATG5-ATG16L complex of approximately 800 kDa (Mizushima et al, 2003).
Atg8 conjugation system
Rather than conjugating to another protein, the Ubl protein Atg8 is attached to phosphatidylethanolamine (PE). The C-terminal Arg 117 residue of Atg8 is initially proteolytically removed by a cysteine protease, Atg4, to expose Gly 116 (Kirisako et al, 2000). This exposed glycine forms a thioester bond with Cys 507 of Atg7, which is also the site that participates in the Atg12–Atg5 conjugation (Ichimura et al, 2000). This feature differentiates Atg7 from most other E1 enzymes, which activate single Ubl proteins. Activated Atg8 is then transferred to the E2-like enzyme Atg3, also through a thioester bond (Ichimura et al, 2000). In the final step of Atg8 lipidation, Gly 116 of Atg8 is conjugated to PE through an amide bond (Ichimura et al, 2000; Fig 2C); Atg8–PE exists in a tightly membrane-associated form.
Although Atg3 shares little sequence homology with other E2 enzymes, structural comparison shows that the head moiety of Atg3 is similar to that of canonical E2 enzymes (Yamada et al, 2007). The amino-acid sequence around the active-site cysteine residue of Atg3 (Cys 234) is also homologous to the corresponding region (Cys 133) in Atg10. Unlike Atg12–Atg5 conjugation, lipidation of Atg8 is reversible; Atg8–PE can be cleaved by Atg4 to release free Atg8 (Kirisako et al, 2000). The Atg8 conjugation cycle, including the second cleavage by Atg4, is important for autophagy; when the processed form Atg8ΔR—which bypasses the need for the initial processing by Atg4—is expressed in an _atg4_Δ mutant, the efficiency of autophagy is low although Atg8–PE can still be formed. Another difference between the Atg8 and Atg12 systems is the regulation of Atg8–PE formation. In yeast, most Atg8 exists in the unconjugated form under nutrient-rich conditions. When autophagy is induced, most Atg8 is converted to the PE-conjugated form (Huang et al, 2000).
In mammalian cells, several homologues of yeast Atg8 have been identified: LC3, GATE16, GABARAP and ATG8L. All of these undergo a modification process similar to that of their yeast counterpart, which is also catalysed by ATG4, ATG3 and ATG7 (Kabeya et al, 2004; Tanida et al, 2003; Tanida et al, 2006; Tanida et al, 2002; Tanida et al, 2001). Among them, LC3 has been best characterized as an autophagosome marker in mammalian cells. LC3 is synthesized as proLC3, and ATG4B processes this precursor into LC3-I with an exposed C-terminal glycine (Kabeya et al, 2004). Catalysed by ATG7 and ATG3, cytosolic LC3-I is transformed to a membrane-bound form, LC3-II, which corresponds to Atg8–PE in yeast. Further analysis shows that LC3 is also attached to PE, and the conjugate can be cleaved by ATG4B. In mammalian cells, the formation of LC3-II can be induced by nutrient depletion or in response to hormone (Kabeya et al, 2000), although the induction level is usually cell line-dependent and tissue-dependent (Mizushima et al, 2004).
Crosstalk between the two conjugation systems
Until recently, E3 activity did not seem to be required for Atg8 lipidation; however, it has now been reported that the Atg12–Atg5 conjugate has an E3-like activity for Atg8 lipidation (Fujita et al, 2008; Hanada et al, 2007). Canonical E3 enzymes bind with both the substrate and the E2 enzyme, and, most importantly, the E3 enzyme stimulates the transfer of ubiquitin from the E2-ubiquitin intermediate to the final target. Corresponding to these criteria, Atg12 interacts with Atg3 in both yeast and mammals (Hanada et al, 2007; Tanida et al, 2002); Atg12–Atg5 also associates with PE-containing liposomes (Hanada et al, 2007); Atg12–Atg5 accelerates the transfer of Atg8 from Atg3 to PE (Hanada et al, 2007); and few Atg8–PE/LC3-II molecules can be detected by Western blot in mutants affecting the Atg12–Atg5-Atg16 complex (Mizushima et al, 2001; Suzuki et al, 2001). However, although Atg12–Atg5 fits well with the E3 criteria, it lacks the domains that are conserved in other known E3 ligases such as HECT-type and RING-type E3 ligases.
Atg12–Atg5-Atg16 is also required for the correct localization of Atg8/LC3. In yeast, GFP-tagged Atg5, Atg16 and Atg8 have a specific localization at the PAS, as well as a cytosolic pool, as shown by fluorescence microscopy (Suzuki et al, 2001). In mammalian cells, ATG12–ATG5-ATG16L also partly co-localizes with LC3 (Mizushima et al, 2003; Mizushima et al, 2001). In yeast, Atg8 does not localize to the PAS in the absence of Atg12–Atg5-Atg16 (Suzuki et al, 2007) and, in mammalian cells, ectopic expression of a plasma membrane-targeted ATG16L directs ATG12–ATG5, as well as LC3, to the plasma membrane (Fujita et al, 2008).
Function of conjugation systems in autophagy
The formation of the Atg12–Atg5 complex and Atg8–PE is essential for autophagy. Mutation of any member of the conjugates, or of other components required for their formation, results in a defect in autophagosome formation (Xie & Klionsky, 2007). Electron-microscopy analysis shows that defects in the recruitment of Atg12–Atg5 or Atg8 to the expanding phagophore do not affect its initiation, suggesting that the two conjugation systems participate in the membrane-expansion step (Mizushima et al, 2001). In yeast, Atg8 is localized to the PAS in an Atg12–Atg5-Atg16-dependent manner, whereas Atg12–Atg5-Atg16 appears at the PAS normally in an _atg8_Δ mutant (Suzuki et al, 2007), implying that the Atg12–Atg5-Atg16 complex functions epistatically with respect to Atg8 in terms of PAS organization.
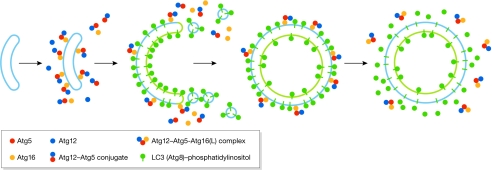
Atg12–Atg5-Atg16 is localized on the expanding phagophore—mostly on the outer surface—and dissociates from the membrane immediately before or after autophagosome completion (Mizushima et al, 2003; Mizushima et al, 2001). On the basis of these phenomena, Atg12–Atg5-Atg16 was proposed as a candidate for a coatomer involved in autophagosome formation. However, a recent quantitative study shows that the number of Atg12–Atg5-Atg16 molecules at the PAS is not sufficient to coat the forming vesicle (Geng et al, 2008). Therefore, promoting Atg8 lipidation and correct localization (Fujita et al, 2008) is the only function identified so far for Atg12–Atg5-Atg16.
As Atg12–Atg5-Atg16 might not be directly involved in membrane expansion, Atg8–PE is the only candidate scaffold for membrane expansion in the two conjugation systems. The amount of Atg8 at the PAS is almost one order of magnitude higher than that of Atg16 (Geng et al, 2008). In contrast to Atg5, Atg8/LC3 is distributed symmetrically on both sides of the phagophore, and part of the population of Atg8/LC3 on the concave surface remains inside the completed autophagosome following completion (Huang et al, 2000; Kabeya et al, 2000; Kirisako et al, 1999). Accumulating evidence indicates a quantitative relationship between the amount of Atg8 and the vesicle size. In yeast, the induction of autophagy results in the upregulation of Atg8 expression (Huang et al, 2000) and the formation of larger autophagosomes (Xie et al, 2008). In mammals, Group A Streptococcus invasion results in the formation of bacteria-containing autophagosomes that are bigger than those induced by starvation and the LC3 expression level is also higher (Nakagawa et al, 2004). These results suggest a possible role of Atg8 as a component of the scaffold that supports the expanding membrane (Fig 3).
Figure 3.
Atg12 and Atg8 conjugation systems in vesicle formation. The initiation of the early-stage phagophore is independent of the presence of the Atg12–Atg5 conjugate, and the Atg12–Atg5-Atg16 complex is subsequently recruited to this structure. Atg12–Atg5-Atg16 redistributes mostly to the outer surface of the phagophore and directs additional Atg8 to this site. The presence of Atg8 on the phagophore supports its expansion. On completion of the autophagosome, the Atg12–Atg5-Atg16 complex dissociates from the vesicle, and Atg4 proteolytically releases the Atg8 that is present on the external surface. Atg, autophagy-related; LC3, microtubule-associated protein 1 light chain 3.
In some specific types of autophagy, Atg8 also functions in cargo selection by interacting with receptors. For example, in the Cvt pathway, Atg8 interacts with Atg19, which is the receptor for precursor Ape1 (Scott et al, 2001; Shintani et al, 2002). In the autophagic degradation of ubiquitinated protein aggregates in mammalian cells, LC3 interacts with p62, which is a ubiquitin-binding protein (Pankiv et al, 2007). These interactions might contribute to the specificity of the processes, so that the vesicle contains only its cargo and excludes other cytosolic components or organelles. Recently, Nakatogawa and colleagues reported that Atg8–PE can mediate membrane tethering and hemifusion in vitro (Nakatogawa et al, 2007), although it is unclear how membrane fusion in vivo occurs only on the edge of the expanding phagophore if Atg8 is distributed evenly on the membrane surface.
Although we have some knowledge regarding the role of Ubls in autophagy, many questions remain to be answered (Sidebar A). Further studies on the mechanism of sequestering vesicle formation are needed to answer them and to elucidate the functions of post-translational modifications in the regulation of this process.
Sidebar A | In need of answers.
- What is the function of Atg8/LC3 and what role does it have in phagophore expansion?
- Do coat elements participate in sequestering vesicle formation?
- How are the different sizes and curvatures of the sequestering vesicles determined?
- What regulates the activity of Atg4 and controls the cleavage of Atg8/LC3–PE?
- What is the nature and purpose of the interaction between Atg8 and the Atg19 cytoplasm-to-vacuole targeting pathway cargo receptor?
- What controls the timing of Atg12–Atg5-Atg16 association with, or dissociation from, the phagophore or autophagosome?
- What are the roles of the mammalian Atg8 homologues?

Jiefei Geng

Daniel J. Klionsky
Acknowledgments
This work was supported by National Institutes of Health Public Health Service grant GM53396 to D.J.K.
References
- Fujita N, Itoh T, Fukuda M, Noda T, Yoshimori T (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19: 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Baba M, Nair U, Klionsky DJ (2008) Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol 182: 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohusmi Y (2007) The Atg12–Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282: 37298–37302 [DOI] [PubMed] [Google Scholar]
- Huang J, Klionsky DJ (2007) Autophagy and human disease. Cell Cycle 6: 1837–1849 [DOI] [PubMed] [Google Scholar]
- Huang W-P, Scott SV, Kim J, Klionsky DJ (2000) The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem 275: 5845–5851 [DOI] [PubMed] [Google Scholar]
- Ichimura Y et al. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 117: 2805–2812 [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ (2007) Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 462: 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang W-P, Stromhaug PE, Klionsky DJ (2002) Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem 277: 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ et al. (2003) A unified nomenclature for yeast autophagy-related genes. Dev Cell 5: 539–545 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Tanida I, Ueno T, Ohsumi M, Ohsumi Y, Kominami E (2001) The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1-E2 complex formation. J Biol Chem 276: 9846–9854 [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N, Ishihara N, Ohsumi Y (2002) Formation of the approximately 350-kDa Apg12–Apg5•Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem 277: 18619–18625 [DOI] [PubMed] [Google Scholar]
- Lupyan D, Leo-Macias A, Ortiz AR (2005) A new progressive-iterative algorithm for multiple structure alignment. Bioinformatics 21: 3255–3263 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y (1998a) A protein conjugation system essential for autophagy. Nature 395: 395–398 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y (1998b) A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem 273: 33889–33892 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Ohsumi Y (1999) Apg16p is required for the function of the Apg12p–Apg5p conjugate in the yeast autophagy pathway. EMBO J 18: 3888–3896 [DOI] [PMC free article] [PubMed]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T (2001) Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 152: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y (2002) Mouse Apg10 as an Apg12-conjugating enzyme: analysis by the conjugation-mediated yeast two-hybrid method. FEBS Lett 532: 450–454 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T (2003) Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12–Apg5 conjugate. J Cell Sci 116: 1679–1688 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Klionsky DJ (2005) Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem 280: 41785–41788 [DOI] [PubMed] [Google Scholar]
- Nakagawa I et al. (2004) Autophagy defends cells against invading group A Streptococcus. Science 306: 1037–1040 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130: 165–178 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Sakai Y, Oku M, van der Klei IJ, Kiel JAKW (2006) Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta 1763: 1767–1775 [DOI] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ (2001) Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell 7: 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ (2004) Autophagy in health and disease: a double-edged sword. Science 306: 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y (1999) Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J 18: 5234–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ (2002) Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell 3: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F (2004) The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 9: 611–618 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 20: 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12: 209–218 [DOI] [PubMed] [Google Scholar]
- Suzuki NN, Yoshimoto K, Fujioka Y, Ohsumi Y, Inagaki F (2005) The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy 1: 119–126 [DOI] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E (1999) Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell 10: 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E (2002) Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem 277: 13739–13744 [DOI] [PubMed] [Google Scholar]
- Tanida I, Komatsu M, Ueno T, Kominami E (2003) GATE-16 and GABARAP are authentic modifiers mediated by Apg7 and Apg3. Biochem Biophys Res Commun 300: 637–644 [DOI] [PubMed] [Google Scholar]
- Tanida I, Sou YS, Minematsu-Ikeguchi N, Ueno T, Kominami E (2006) Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. FEBS J 273: 2553–2562 [DOI] [PubMed]
- Tanida I, Tanida-Miyake E, Ueno T, Kominami E (2001) The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem 276: 1701–1706 [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S, Bugg CE, Cook WJ (1987) Structure of ubiquitin refined at 1.8 Å resolution. J Mol Biol 194: 531–544 [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky DJ (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19: 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Suzuki NN, Hanada T, Ichimura Y, Kumeta H, Fujioka Y, Ohsumi Y, Inagaki F (2007) The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J Biol Chem 282: 8036–8043 [DOI] [PubMed] [Google Scholar]
- Young ARJ, Chan EYW, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119: 3888–3900 [DOI] [PubMed] [Google Scholar]