Guidelines for Naming Nonprimate APOBEC3 Genes and Proteins (original) (raw)
APOBEC3 GENES ARE UNIQUE TO MAMMALS, BUT COPY NUMBERS VARY SIGNIFICANTLY
APOBEC3 (A3) proteins are of considerable interest because most are potent DNA cytidine deaminases that have the capacity to restrict the replication and/or edit the sequences of a wide variety of parasitic elements, including many retroviruses and retrotransposons (reviewed in references 5, 8-10, and 14). Likely substrates include (i) lentiviruses, such as human immunodeficiency virus type 1, human immunodeficiency virus type 2, simian immunodeficiency virus, maedi-visna virus, feline immunodeficiency virus, and equine infectious anemia virus; (ii) alpha-, beta-, gamma-, and deltaretroviruses, such as Rous sarcoma virus, Mason-Pfizer monkey virus or mouse mammary tumor virus, murine leukemia virus or feline leukemia virus, and human T-cell leukemia virus or bovine leukemia virus, respectively; (iii) spumaviruses, such as primate foamy virus and feline foamy virus; (iv) hepadnaviruses, such as hepatitis B virus; (v) endogenous retroviruses and long terminal repeat retrotransposons, such as human endogenous retrovirus K, murine intracisternal A particle, murine MusD, and porcine endogenous retrovirus; (vi) non-long terminal repeat retroposons, such as L1 and Alu; and (vii) DNA viruses, such as adeno-associated virus and human papillomavirus. Over the past few years, there has also been an increasing appreciation for the multiple, distinct mechanisms that parasitic elements use to coexist with the A3 proteins of their hosts. Together, these observations indicate that the evolution of the A3 proteins has been driven by a requirement to minimize the spread of exogenous and endogenous genetic threats. The likelihood that the A3 proteins might exist solely for this purpose has been supported recently by studies indicating that A3-deficient mice have no obvious phenotypes apart from a notable increase in susceptibility to retrovirus infection (16, 19, 21, 23).
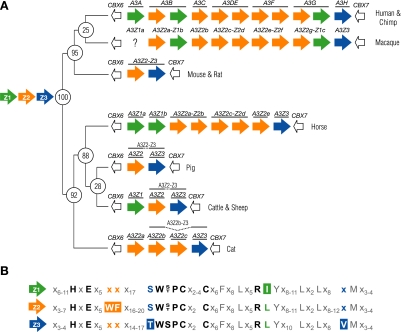
A3 genes are specific to mammals and are organized in a tandem array between two vertebrate-conserved flanking genes, CBX6 and CBX7 (Fig. 1A) (e.g., see reference 13). Based on a limited number of genomic sequences, it is already clear that the A3 copy number can vary greatly from mammal to mammal. For instance, mice have one A3 gene (10, 16), pigs have two (13), cattle and sheep have three (13), cats have four (17), horses have six (2), and humans and chimpanzees have seven (4, 10, 11). Other mammals are likely to have copy numbers within this range, but the cat and horse loci, in particular, highlight the difficulty in making such predictions (2, 17).
FIG. 1.
(A) Schematics of the A3 repertoires of mammals whose genomes have been sequenced. Z1, Z2, and Z3 domains are shown in green, orange and blue, respectively. For all of the indicated species (and likely all mammals), CBX6 is located immediately upstream and CBX7 downstream of the A3 locus. Either macaque A3A does not exist, or its genomic sequence is not quite complete. The inferred ancestral A3 repertoire was deduced through comparative studies (13). The numbers at the phylogenetic tree branch points indicate the approximate time, in millions of years, since the divergence of the ancestors of the clades of the indicated present-day species (1). (B) Highlights of amino acid conservation among the three distinct Z-domain groups and within each individual group (based on multiple sequence alignments) (13). Residues discussed in the text are in color or boldface, and other notable residues are in gray. An “x” specifies nearly any amino acid.
EACH APOBEC3 GENE IS COMPRISED OF ONE OR TWO ZINC-COORDINATING DOMAINS
Naming the mammalian A3 genes is complicated further by the fact that each gene encodes a single- or a double-zinc (Z)-coordinating-domain protein. For instance, human A3A, A3C, and A3H encode single-Z-domain proteins, whereas human A3B, A3DE, A3F, and A3G encode double-Z-domain proteins. The Z domain is required for catalytic activity, but some domains have not elicited activity and can therefore be regarded as pseudocatalytic. Nevertheless, all Z domains can be readily identified by four invariant residues, namely, one histidine, one glutamate, and two cysteines, organized Hx1Ex23-28Cx2-4C (x can be nearly any 1 of the 20 amino acids, and underlining indicates the invariant residues) (Fig. 1B and see below). The histidine and two cysteines are required to bind a single zinc atom and, at least for catalytic domains, the glutamate is predicted to promote the formation of a hydroxide ion required for deamination.
Each Z domain clearly belongs to one of three distinct phylogenetic clusters, originally termed Z1b, Z1a, and Z2 (7; adopted in references 6, 18, and 20). However, while we ac-knowledge the logical nature of these Z-based groupings, we propose a simplification of the scheme to Z1, Z2, and Z3, respectively. This minor nomenclature change was motivated because (i) lowercase letters are needed to help describe unique A3 variants (see below), (ii) a key mammalian ancestor likely had a CBX6-Z1-Z2-Z3-CBX7 locus organization (13), and (iii) the Z3 domain has so far been found to be invariably located at the distal end of the locus, next to CBX7 (Fig. 1A).
Z-domain assignments can be made simply by scanning predicted polypeptide sequences for key identifying residues (Fig. 1B). This determination is facilitated by the fact that the Z domain of all known A3 genes is encoded by a single exon. For instance, Z1 domains have a unique isoleucine (I) adjacent to a conserved arginine common to all DNA deaminases (3). Z2 domains possess a unique tryptophan-phenylalanine (WF) motif five residues after the (pseudo)catalytic glutamate. Finally, Z3 domains have a TWSPCx2-4C zinc-coordinating motif, whereas both the Z1 and Z2 domains have a SWS/TPCx2-4C motif. Since many A3 proteins have been subject to positive selection (22), this Z-based scheme is also substantially more robust to evolutionary constraints and pressures that have acted (and continue to act) on A3 proteins in different lineages.
However, although these simple rules enable initial Z-domain assignments, it should be noted that several other differences combine to distinguish each of the three Z types, and final assignments should be verified by comprehensive phylogenetic analyses. One should also be aware of the fact that the mammalian A3 locus is frequently involved in genetic recombination events, such as unequal crossing-over events (leading to deletions or insertions) and gene conversions (e.g., see reference 13). Thus, to minimize the potentially confounding effects of recombination, we further recommend (at least for the purposes of nomenclature) that A3 gene descriptions be based exclusively on Z-domain assignments (i.e., based on phylogenetic analyses of the Z-domain-encoding exon) (e.g., see Fig. 1A and reference 13).
Z-DOMAIN-BASED NOMENCLATURE SYSTEM FOR NONPRIMATE APOBEC3 GENES
With new technologies delivering tidal waves of genomic and transcribed sequences to the scientific community, it is important to have nomenclature systems in place to facilitate the annotation, dissemination, and comparison of specific genes and gene families. The current Human Genome Organization conventions suggest that the human gene name be used to annotate the orthologous genes of nonhuman species (http://www.genenames.org). The Human Genome Organization system can be applied readily to the A3 genes of primates such as the chimpanzee and the rhesus macaque, which align nearly domain-for-domain with the human A3 locus (Fig. 1A). However, the A3 loci of nonprimate mammals pose a particularly difficult problem, because they vary in size, Z-domain type, and Z-domain organization. Read-through transcription, alternative splicing, and internal transcription initiation further complicate naming schemes (e.g., see references 13 and 17). Most importantly, it is impossible (and incorrect) to deduce orthologous relationships between humans and nonprimate mammals, because each species' A3 proteins are the product of a unique, divergent evolutionary history that was shaped by immeasurable selective pressures.
Therefore, to simplify matters, we propose the following Z-domain-based nomenclature system that can be applied easily to annotate and describe the APOBEC3 repertoire of any nonprimate mammal. It is based on the fact that the A3 genes are clearly modular in nature, consisting of one Z domain (Z1, Z2, or Z3) or some combination of two Z domains (Z2-Z1, Z2-Z2, or Z2-Z3) (2, 13, 17). Other combinations may very well exist, but they have yet to be described. This Z-domain-based system is best applied once a species' entire A3 genomic locus has been determined, and it does not require immediate knowledge of mRNA or protein-coding capacity.
First, once an A3 locus has been sequenced (ideally, completely), the Z-domain type should be assigned as described above. A simple example is the A3 locus in cattle, which consists of three distinct Z domains in a Z1-Z2-Z3 organization (13). A more complex example is that of the horse, which consists of two Z1 domains, five Z2 domains, and a single Z3 domain (2). Second, in such an instance when multiple domains of a single Z type exist, we propose that lowercase letters be used to distinguish each distinct domain (ideally applied starting at the CBX6 side of the locus and ending at the CBX7 side, i.e., starting at the 5′ end). For instance, the eight-Z-domain horse A3 repertoire would be designated Z1a-Z1b-Z2a-Z2b-Z2c-Z2d-Z2e-Z3. Finally, based on mRNA expression data, which will undoubtedly reveal how the Z domains mix and match in vivo, additional assignments can be made. Single-Z-domain genes, mRNAs, and proteins can be annotated simply by adding the APOBEC3 (A3) prefix. For instance, cattle have three APOBEC3 genes: A3Z1, A3Z2, and A3Z3 (13). Following this logic, double-Z-domain genes, mRNAs, and proteins can be annotated by adding the A3 prefix and pairing the Z-domain designations. For instance, cattle also have an A3Z2-Z3 protein (13), and the coding potential of the horse A3 repertoire can be described as A3Z1a, A3Z1b, A3Z2a-Z2b, A3Z2c-Z2d, A3Z2e, and A3Z3 (e.g., see reference 2 and Fig. 1A). New names for all of the A3 genes of nonprimate mammals whose A3 genomic loci are “complete” are listed in Table 1.
TABLE 1.
APOBEC3 genes and proteins of representative nonprimate mammals
| Genus and species (common name) | Old name (reference) | New name (reference) | ||
|---|---|---|---|---|
| Genea | Proteina | Geneb | Protein | |
| Bos taurus (cattle) | A3Z1 (13) | A3Z1 | ||
| A3Z2 (13) | A3Z2 | |||
| A3Z3 (13) | A3Z3 | |||
| A3F (12) | A3F | A3Z2-Z3 (13) | ||
| Equus caballus (horse) | A3A1 (2) | A3A1 | A3Z1a | A3Z1a |
| A3A2 (2) | A3A2 | A3Z1b | A3Z1b | |
| A3F1 (2) | A3F1 | A3Z2a-Z2b | A3Z2a-Z2b | |
| A3F2 (2) | A3F2 | A3Z2c-Z2d | A3Z2c-Z2d | |
| A3C (2) | A3C | A3Z2e | A3Z2e | |
| A3H (2) | A3H | A3Z3 | A3Z3 | |
| Felis catus (cat) | A3Cc (17) | A3Cc | A3Z2a | A3Z2a |
| A3Ca (17) | A3Ca | A3Z2b | A3Z2b | |
| A3Cb (17) | A3Cb | A3Z2c | A3Z2c | |
| A3H (17) | A3H | A3Z3 | A3Z3 | |
| A3CH (17) | A3Z2b-Z3 | |||
| Mus musculus (mouse) | A3 (15) | A3 | A3Z2-Z3 | A3Z2-Z3 |
| Ovis aries (sheep) | A3Z1 (13) | A3Z1 | ||
| A3Z2 (13) | A3Z2 | |||
| A3Z3 (13) | A3Z3 | |||
| A3F (12) | A3F | A3Z2-Z3 (13) | ||
| Rattus norvegicus (rat) | A3 | A3 | A3Z2-Z3 | A3Z2-Z3 |
| Sus scrofa (pig) | A3Z2 (13) | A3Z2 | ||
| A3Z3 (13) | A3Z3 | |||
| A3F (12) | A3F | A3Z2-Z3 (13) |
At first glance, this new nomenclature system may appear cumbersome. However, we suspect that continual exposure and practice will yield both familiarity and, possibly, a colloquial “short form” that lacks common denominators. Again, using cattle and horses as examples, the former have Z1, Z2, Z3, and Z2-3 types of A3 proteins, and the latter have Z1a, Z1b, Z2ab, Z2cd, Z2e, and Z3 types of A3 proteins.
It also is worth mentioning that a Z-domain-based system is also possible for the primate A3s (Fig. 1A). A complete conversion to this system would certainly facilitate intra-Z-type and interspecies comparisons, but we fully recognize that the well-established (and popular) human A3A through A3H designations are not likely to be superseded (Fig. 1A). We further recognize that the mouse may also be a special case, because the generic A3 designation has already been used to describe its single (albeit double-Z-domain) gene. However, regardless of whether the new nomenclature scheme is adopted, it is important to emphasize again that it guards against the false implication of orthology between certain human A3 genes and the A3 genes found in other mammals. Previously, A3 genes have been tentatively named on the basis of BLAST score matches, which have been shown to be a notoriously poor means of establishing orthology, especially when reciprocal best BLAST hits are not employed. Thus, the new nomenclature scheme not only is simple and logical but also is more formally correct than current schemes.
Finally, it is important to point out that the new system readily accommodates A3 variants created by read-through transcription and alternative splicing. For instance, the feline A3 locus, which encodes four similarly designated single-domain proteins and a novel A3Z2b-Z3 variant (17), can now be designated A3Z2a-A3Z2b-A3Z2c-A3Z3. Moreover, a numeric suffix can be added to each designation to accommodate splice variants. Overall, we hope that the intrinsic logic of the simplified Z-domain-based nomenclature system will enable the mammalian A3 genes to be fully described and appropriately included in a wealth of comparative studies to better understand a broad range of host-pathogen conflicts.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
▿
Published ahead of print on 5 November 2008.
REFERENCES
- 1.Bininda-Emonds, O. R., M. Cardillo, K. E. Jones, R. D. MacPhee, R. M. Beck, R. Grenyer, S. A. Price, R. A. Vos, J. L. Gittleman, and A. Purvis. 2007. The delayed rise of present-day mammals. Nature 446507-512. [DOI] [PubMed] [Google Scholar]
- 2.Bogerd, H. P., R. L. Tallmadge, L. J. Oaks, S. Carpenter, and B. R. Cullen. 2008. Equine infectious anemia virus resists the antiretroviral activity of equine APOBEC3 proteins through a packaging-independent mechanism. J. Virol. 8211889-11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, K. M., E. Harjes, P. J. Gross, A. Fahmy, Y. Lu, K. Shindo, R. S. Harris, and H. Matsuo. 2008. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 452116-119. [DOI] [PubMed] [Google Scholar]
- 4.Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 43769-87. [DOI] [PubMed] [Google Scholar]
- 5.Chiu, Y. L., and W. C. Greene. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26317-353. [DOI] [PubMed] [Google Scholar]
- 6.Conticello, S. G., M. A. Langlois, Z. Yang, and M. S. Neuberger. 2007. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv. Immunol. 9437-73. [DOI] [PubMed] [Google Scholar]
- 7.Conticello, S. G., C. J. Thomas, S. Petersen-Mahrt, and M. S. Neuberger. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22367-377. [DOI] [PubMed] [Google Scholar]
- 8.Cullen, B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 801067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goila-Gaur, R., and K. Strebel. 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4868-877. [DOI] [PubMed] [Google Scholar]
- 11.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79285-296. [DOI] [PubMed] [Google Scholar]
- 12.Jónsson, S. R., G. Haché, M. D. Stenglein, S. C. Fahrenkrug, V. Andrésdóttir, and R. S. Harris. 2006. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 345683-5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaRue, R. S., S. R. Jónsson, K. A. T. Silverstein, M. Lajoie, D. Bertrand, N. El-Mabrouk, I. Hötzel, V. Andresdottir, T. P. L. Smith, and R. S. Harris. 2008. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 9104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malim, M. H., and M. Emerman. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3388-398. [DOI] [PubMed] [Google Scholar]
- 15.Mariani, R., D. Chen, B. Schröfelbauer, F. Navarro, R. König, B. Bollman, C. Münk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 11421-31. [DOI] [PubMed] [Google Scholar]
- 16.Mikl, M. C., I. N. Watt, M. Lu, W. Reik, S. L. Davies, M. S. Neuberger, and C. Rada. 2005. Mice deficient in APOBEC2 and APOBEC3. Mol. Cell. Biol. 257270-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Münk, C., T. Beck, J. Zielonka, A. Hotz-Wagenblatt, S. Chareza, M. Battenberg, J. Thielebein, K. Cichutek, I. G. Bravo, S. J. O'Brien, M. Löchelt, and N. Yuhki. 2008. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 9R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.OhAinle, M., J. A. Kerns, H. S. Malik, and M. Emerman. 2006. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 803853-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okeoma, C. M., N. Lovsin, B. M. Peterlin, and S. R. Ross. 2007. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature 445927-930. [DOI] [PubMed] [Google Scholar]
- 20.Rogozin, I. B., M. K. Basu, I. K. Jordan, Y. I. Pavlov, and E. V. Koonin. 2005. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle 41281-1285. [DOI] [PubMed] [Google Scholar]
- 21.Santiago, M. L., M. Montano, R. Benitez, R. J. Messer, W. Yonemoto, B. Chesebro, K. J. Hasenkrug, and W. C. Greene. 2008. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science 3211343-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawyer, S. L., M. Emerman, and H. S. Malik. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeda, E., S. Tsuji-Kawahara, M. Sakamoto, M. A. Langlois, M. S. Neuberger, C. Rada, and M. Miyazawa. 2008. Mouse APOBEC3 restricts Friend leukemia virus infection and pathogenesis in vivo. J. Virol. 8210998-11008. [DOI] [PMC free article] [PubMed] [Google Scholar]