Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways (original) (raw)
. Author manuscript; available in PMC: 2009 Oct 1.
Summary
Background
Psoriasis vulgaris is an inflammatory skin disease mediated by Th1 and Th17 cytokines, yet the relative contribution of interferon (IFN)-γ, interleukin (IL)-17 and IL-22 on disease pathogenesis is still unknown.
Objectives
In this study, we sought to identify the cytokines produced by skin-resident T cells in normal skin, localize the receptors for these cytokines, and examine how these cytokines alter gene expression profiles of the cells bearing cognate receptors.
Methods
We used intracellular cytokine staining and flow cytometry to evaluate T cell cytokine production, and immunohistochemistry and double-label immunofluorescence to localize cytokine receptors in skin. Gene array analysis of cytokine-treated keratinocytes was performed using moderated paired _t_-test controlling for false discovery rate using the Benjamini–Hochberg procedure.
Results
We demonstrate that T-helper cells producing IL-17, IL-22 and/or IFN-γ, as well as the cells bearing cognate cytokine receptors, are present in normal human skin. Keratinocytes stimulated with IL-17 expressed chemokines that were different from those induced by IFN-γ, probably contributing to the influx of neutrophils, dendritic cells and memory T cells into the psoriatic lesion. In contrast, IL-22 downregulated genes associated with keratinocyte differentiation and caused epidermal alterations in an organotypic skin model.
Conclusions
Our results suggest that the Th17 cytokines IL-17 and IL-22 mediate distinct downstream pathways that contribute to the psoriatic phenotype: IL-17 is more proinflammatory, while IL-22 retards keratinocyte differentiation.
Keywords: interleukin-17, interleukin-22, keratinocytes, Th1, Th17
Psoriasis vulgaris is a common chronic inflammatory skin disease characterized by hyperproliferative epidermis and mixed cutaneous lymphocytic infiltrate. While initially regarded as a primary disease of keratinocyte differentiation, effective immune-modulating therapies demonstrate the vital role played by the immune system in psoriatic disease pathogenesis.1–4 The T cells involved in lesion formation were initially thought to be Th1 differentiated based on interferon (IFN)-γ and interleukin (IL)-2 production.5–7 However, the recent discovery of the Th17 T-helper cell subset, and its potential involvement in psoriasis, generates even more complexity to this disease.
Th17 cells have recently been classified as distinct from Th1 and Th2 subsets.8,9 They are defined by the ability to synthesize IL-17 in response to antigen-presenting cell-derived IL-23 and other differentiating cytokines.10–13 In addition, Th17 cells have been reported to cosynthesize IL-17 and IFN-γ14 as well as IL-22.15,16 Indeed, in murine systems, IL-22 production occurs overwhelmingly within the Th17 subset.15
Psoriatic skin lesions are reported to have increased gene expression of IL-23,17 IL-17 and IL-22,18–21 prompting investigators to probe deeper into the potential involvement of Th17 cells in psoriasis. While models of epidermal hyperproliferation have focused on IL-22 as being central to psoriasis pathogenesis via induction of keratinocyte proliferation and acanthosis,22–24 both IL-22 and IL-17 have been shown to induce keratinocyte gene expression of antimicrobials β-defensin 2, β-defensin 3, S100A8 and S100A9, all upregulated in psoriatic lesions.15,22,25 Besides the increased expression of antimicrobial genes, however, the contribution of IL-17 to psoriasis pathogenesis has not been thoroughly investigated. This is in contrast to IL-17 being extensively implicated in chemokine-induced neutrophil recruitment in asthma, chronic obstructive pulmonary disease and cystic fibrosis.26–28
The chemokines that are considered to be neutrophil chemoattractants belong to the ELR+ CXC subfamily, named by the presence of a Glu-Leu-Arg motif at residues 4–6.29,30 Members of this subfamily include CXCL1–8, except CXCL4.29 Indeed, IL-17 has previously been shown to induce the production of CXCL1 and CXCL8 in bronchial epithelial cells,27,28 fibroblasts31 and keratinocytes,32 and neutralizing antibodies to IL-17 or its receptor can block this induction.28,32
Even with increasing evidence for the involvement of Th17 cells in psoriasis pathogenesis, the relative effects of the Th17 cytokines IL-17 and IL-22 and the Th1 cytokine IFN-γ on the skin are unknown. In this study, we sought to identify the cytokines produced by skin-resident T cells in normal skin, localize the receptors for these cytokines, and examine how these cytokines alter gene expression profiles of the cells bearing cognate receptors.
Materials and methods
Skin samples
Skin punch biopsies (6 mm diameter) were obtained from normal volunteers (n = 5) and patients with moderate-to-severe chronic plaque psoriasis (n = 16) under a Rockefeller University Institutional Review Board-approved protocol. The biopsy specimens were frozen in OTC (Sakura, Torrance, CA, U.S.A.) and stored at −80°C for immunohistochemistry and immunofluorescence and flash frozen in liquid nitrogen for RNA extraction and analysis. Dermal single-cell suspensions were obtained from abdominoplasty by overnight incubation in dispase (Invitrogen, Carlsbad, CA, U.S.A.) and collagenase (Roche, Indianapolis, IN, U.S.A.) 1 mg mL−1 at 4°C, peeling off the epidermis, and culturing the dermis for 36–48 h at 37°C in RPMI 1640 (Gibco, Carlsbad, CA, U.S.A.) supplemented with 10% pooled human serum (Mediatech Inc., Manassas, VA, U.S.A.), 0.1% gentamicin reagent solution (Gibco) and 1% 1 mol L−1 HEPES buffer (Sigma, St Louis, MO, U.S.A.). Epidermal single-cell suspensions were obtained by incubation in 0.25% trypsin/ethylenediamine tetraacetic acid (EDTA) (Gibco) for 10 min at 37°C, then in RPMI 1640 with 10% pooled human serum, 0.1% gentamicin reagent solution and 1% 1 mol L−1 HEPES buffer overnight.
Peripheral blood samples
Peripheral blood mononuclear cells from freshly drawn blood of normal volunteers (n = 3) were purified by gradient centrifugation with Ficoll-Paque Plus (Pharmacia, Piscataway, NJ, U.S.A.), collected at the interface, then washed with phosphate-buffered saline (PBS) prior to fluorescence-activated cell sorting (FACS) analysis.
Primary keratinocyte cultures
Primary pooled human keratinocytes (n = 3) were obtained from Yale Skin Diseases Research Center core facility and cultured in RPMI 1640 with 10% pooled human serum, 0.1% gentamicin reagent solution and 1% 1 mol L−1 HEPES buffer at 37°C as above. Once 80% confluent, the medium was supplemented with or without the cytokines recombinant human (rh)-IL-17 (R&D Systems, Minneapolis, MN, U.S.A.) 200 ng mL−1, rh-IL-22 (Peprotech Inc., Rocky Hill, NJ, U.S.A.) 200 ng mL−1 or rh-IFN-γ (R&D Systems) 20 ng mL−1 for 24 h before harvesting for other analyses.
Human full-thickness skin model
Full-thickness human skin models (MatTek Corp., Ashland, MA, U.S.A.) were incubated in assay media (MatTek Corp.) supplemented with or without the cytokines rh-IL-17 200 ng mL−1 or rh-IL-22 200 ng mL−1 for 4 days (n = 3). Media, with or without supplemention, were changed every 48 h. On days 2 and 4, the skin models were harvested for histological and RNA analyses.
Antibodies
All antibodies used for immunofluorescence and FACS are listed in Table 1 and Table 2.
Table 1.
Antibodies used for immunohistochemistry and immunofluorescence
| Antigen | Manufacturer | Clonea | Isotype | Dilution | Amplification/detectionb |
|---|---|---|---|---|---|
| CD11c | BDPharmingen,San Jose, CA,U.S.A. | B-ly6 | IgG1 | 1:100 | Goat antimouse IgG1(A568/A488) |
| IL-17R | Amgen,ThousandOaks, CA,U.S.A. | M202 | IgG2a | 1:100 | Goat antimouse IgG2a(A488) |
| IL-22R | R&DSystems,Minneapolis,MN, U.S.A. | 305405 | IgG1 | 1:100 | Goat antimouse IgG1(A488) |
| CXCR1 | eBioscience,San Diego,CA, U.S.A. | 8F1-1-4 | IgG2b | 1:100 | - |
| CXCR2 | BDPharmingen,San Jose, CA,U.S.A | 6C6 | IgG1 | 1:100 | - |
| Vimentin | Neomarkers,Fremont, CA,U.S.A. | V9 | IgG1 | 1:100 | Goat antimouse IgG1(A568) |
Table 2.
Antibodies used for flow cytometry
| Antigen–fluorophore | Manufacturer | Clonea | Isotype | Dilution |
|---|---|---|---|---|
| IL-22 | R&D Systems,Minneapolis, MN,U.S.A. | 142928 | IgG1 | 1:33 |
| CD3–Pacific Blue | eBioscience, SanDiego, CA, U.S.A. | OKT3 | IgG2a | 1:33 |
| CD4–PE–Cy7 | eBioscience, SanDiego, CA, U.S.A. | RPA-T4 | IgG1 | 1:33 |
| CD8–PerCP–Cy5.5 | BD Pharmingen,San Jose, CA,U.S.A | SK1 | IgG1 | 1:20 |
| IL-17–Alexa 488 | eBioscience, SanDiego, CA, U.S.A. | eBio64DEC17 | rat IgG2a | 1:20 |
| IFN-γ–Alexa 700 | BD Pharmingen,San Jose, CA,U.S.A. | B27 | IgG1 | 1:33 |
| Live-Dead–aqua marina | Invitrogen, Carlbad,CA, U.S.A. | NA | NA | 1:100 |
| CD11c–PE | BD Pharmingen,San Jose, CA,U.S.A. | S-HCL-3 | IgG2b | 1:33 |
| CD45–PerCP | BD Pharmingen,San Jose, CA,U.S.A. | 2D1 | IgG1 | 1:20 |
| IL-17R | Amgen, ThousandOaks, CA, U.S.A. | M202 | IgG2a | 1:33 |
| IL-22R | R&D Systems,Minneapolis, MN,U.S.A. | 305405 | IgG1 | 1:33 |
Immunohistochemistry
Tissue sections were stained with haematoxylin (Fisher Scientific, Pittsburgh, PA, U.S.A.) and eosin (Shandon, Pittsburg, PA, U.S.A.), or with purified mouse antihuman monoclonal antibodies listed in Table 1. Biotin-labelled horse antimouse antibodies (Vector Laboratories, Burlingame, CA, U.S.A.) were amplified with avidin-biotin complex (Vector Laboratories) and developed with chromogen 3-amino-9-ethylcarbazole (Sigma Aldrich, St Louis, MO, U.S.A.). Appropriate negative controls were used.
Immunofluorescence
Skin sections were stained as previously described19 using antibodies listed in Table 1. Images were acquired using appropriate filters of a Zeiss Axioplan 2I microscope with Plan Apochromat 20 × 0.7 numerical aperture lens and a Hagamatsu orca ER-cooled charge-coupled device camera, controlled by METAVUE software (Molecular Devices, Sunnyvale, CA, U.S.A.).
Reverse transcriptase–polymerase chain reaction
RNA was extracted from primary human keratinocytes, full-thickness skin equivalents and human skin using the RNeasy Mini Kit (Qiagen, Valencia, CA, U.S.A.). Reverse transcriptase (RT)–polymerase chain reaction (PCR) was performed using EZ PCR core reagents, primers and probes (Applied Biosystems, Foster City, CA, U.S.A.) as previously published.33 Sequences of primers and probes used in this study were as follows: CCL20 (Hs00171125_m1), CXCL1 (Hs00236937_m1), CXCL3 (Hs00171061_m1), CXCL5 (Hs00171085_m1), CXCL6 (Hs00237017_m1), CXCL8 (Hs00174103_m1), S100A7 (HS00161488_m1) and DEFB4 (Hs00175474_m1). The data were analysed and samples quantified by software provided with Applied Biosystems PRISM 7700 (Sequence Detection Systems, ver. 1.7). Data were normalized to HARP housekeeping gene.
Gene array
RNA was extracted using the RNeasy Mini Kit (Qiagen), and DNA was removed with on-column DNAse digestion using RNAse-free DNAse Set (Qiagen), and used for either RT-PCR or gene array. For each Affymetrix genechip, 4 µg total RNA was reverse transcribed, amplified, and labelled as described previously using BioArray High Yield RNA Transcription Labeling Kit (Enzo Biochem Inc., Farmingdale, NY, U.S.A.).34 Fifteen micrograms of the biotinylated cRNA was then hybridized to Affymetrix Human Genome U133A 2.0 Array (14 500 probe sets) (Affymetrix, Santa Clara, CA, U.S.A.). The chips were washed, stained with streptavidin-phycoerythin, and scanned with an Hewlett-Packard HP GeneArray Scanner (Hewlett-Packard, Palo Alto, CA, U.S.A.).
Preprocessing and statistical analysis was conducted in R (http://www.rproject.org/). GeneChip CEL files were scrutinized for spatial artifacts using Harshlight package (http://asterion.rockefeller.edu/Harshlight/index2.html).35 Row intensities values (CEL files) were preprocessed to obtained expression values using GCRMA algorithm.
Fluorescence-activated cell sorting analysis
Cells were stained with the antibodies listed in Table 2. Briefly, cells were stained for cell surface molecules for 20 min at 4°C, washed with FACSwash (PBS, 0.1% sodium azide and 2% fetal bovine serum) (BD Biosciences, San Jose, CA, U.S.A.) and resuspended in FACSwash (BD Biosciences). For intracellular cytokine staining assays, cells were activated for 4 h using 25 ng mL−1 phorbol myristate acetate and 2 µg mL−1 ionomycin, in the presence of 10 µg mL−1 brefeldin A (all Sigma Aldrich) at 37°C. Unactivated controls were treated with brefeldin A only. EDTA 2 mmol L−1 (Fisher Scientific) was added for 10 min at 37°C to stop activation. Cells were then incubated in aqua marina live/dead dye (Invitrogen) for 30 min for dead cell discrimination then fixed with 4% paraformaldehyde (BD Biosciences) for 20 min. The cells were washed, blocked in 1:100 mouse serum (BD Biosciences), permeabilized in FACSPerm (BD Biosciences), incubated for 30 min with fluorochrome-conjugated monoclonal antibodies to cell surface molecules and intracellular cytokines, washed, and collected. Samples were acquired by an LSR-II flow cytometer (BD Biosciences) and analysed with FlowJo software (Treestar, Ashland, OR, U.S.A.). Keratinocyte single-cell suspensions were analysed after gating out CD45+ and CD11c+ cells. All incubation steps were done on ice. Appropriate isotype controls were used.
Statistical analysis
Two-tailed paired _t_-test was used to compare psoriasis nonlesional and lesional RT-PCR data. Two-tailed Student’s _t_-test was used to analyse RT-PCR data between keratinocyte treatment groups, and between full-thickness skin models. The two-tailed _P_-values are designated as P < 0.05 (*), _P_ < 0.01 (**) and _P_ < 0.005 (***). For gene array analysis, probe sets with SD > 0.1 and at least one sample with expression > 2 were included. Moderated paired _t_-test available at limma package from R was performed. The resulting _P_-value was adjusted for multiple hypotheses testing, controlling the false discovery rate (FDR) using the Benjamini–Hochberg procedure. Genes with FDR < 0.1 and more than 1.5-fold change were considered significant.
Results
Distinct populations of interleukin (IL)-17 and IL-22-producing T cells are present in normal human dermis and peripheral blood
We have previously shown that there are discrete populations of IL-17 and IFN-γ-producing T-helper cells in psoriatic and normal dermis and peripheral blood.18 As murine Th17 cells can produce both IL-17 and IL-22,15 we wanted to investigate whether IL-17-producing cells in normal human skin and peripheral blood could also produce IL-22.
Using intracellular cytokine staining and eight-colour flow cytometry, we simultaneously analysed the expression of IFN-γ, IL-17 and IL-22 within live CD3+ CD4+ CD8− T cells. We found that approximately 2% of CD4+ T cells in peripheral blood, and 1% in normal dermis, produced IL-17 – defined as Th17 cells (Fig. 1, second and third columns). A larger population of CD4+ T cells, 16% in the blood and 7% in the dermis, produced IFN-γ but not IL-17 (Fig. 1, third column). These cells were defined as Th1 cells.
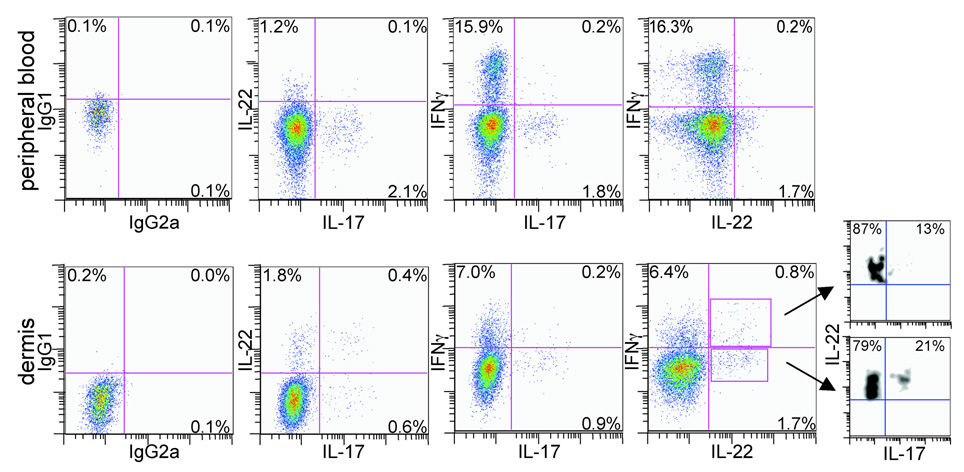
Fig 1.
CD4+ T cells producing interleukin (IL)-17, IL-22 and interferon (IFN)-γ are present in peripheral blood (upper panels) and normal dermis (lower panels). Intracellular cytokine staining and flow cytometric analysis show distinct populations of CD4+ T cells producing IL-17, IL-22 and IFN-γ in peripheral blood. In normal human dermis there are CD4+ T cells producing only IFN-γ, IL-17 or IL-22, and CD4+ T cells that cosynthesize IL-17 and IL-22, IL-17 and IFN-γ, IL-22 and IFN-γ or all three cytokines. Fluorescence-activated cell sorting plots are representative of two separate experiments. Leftmost panels are isotype controls.
Approximately 1.2–1.7% of all CD4+ T cells in peripheral blood produced only IL-22, independent of IFN-γ or IL-17 production (Fig. 1, second and fourth columns). In normal dermis, 1.8% of all CD4+ T cells produced only IL-22, while 0.4% produced both IL-22 and IL-17, and 0.8% produced IL-22 and IFN-γ (Fig. 1, lower panels).
In order to determine the percentage of IL-22-producing cells that also produce IL-17, we gated on IL-22+ IFN-γ+ or IL-22− IFN-γ− cells (Fig. 1, lower right panel) and plotted IL-22 vs. IL-17 expression. We found that while some of the IL-22+ IFN-γ+ cells and IL-22+ IFN-γ− cells produced IL-17 (13% and 21%, respectively), the majority did not (87% and 79%, respectively).
Interleukin (IL)-17 and IL-22 receptors are constitutively expressed by keratinocytes
We assessed expression and localization of receptors for IL-17 (IL-17R) and IL-22 (IL-22R1) on cells in normal human skin using immunohistochemical staining (n = 5). Both IL-17R and IL-22R1 were strongly expressed on cell surfaces of viable keratinocytes throughout the epidermis (Fig. 2a, b; inset is negative control). Keratinocyte surface expression of these receptors was confirmed by flow cytometry on single-cell suspensions of normal epidermis (Fig. 2c, d).
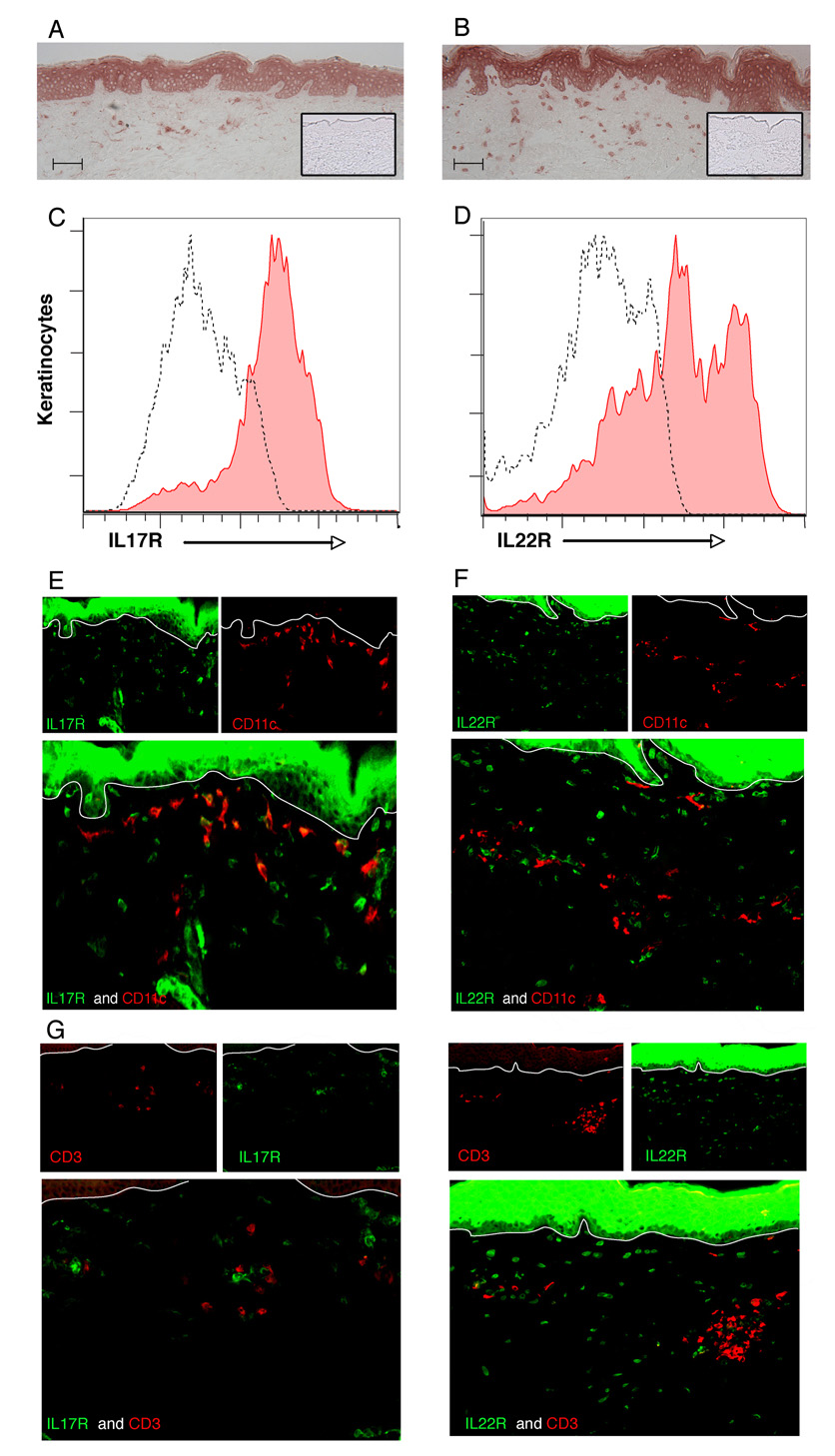
Fig 2.
Interleukin (IL)-17 and IL-22 receptors are expressed by keratinocytes and some dermal cells. Immunohistochemical analysis of IL-17R (a) and IL-22R1 (b) on normal human skin (n = 5) (inset, negative control). Scale bars = 100 µm. Confirmation of surface expression of IL-17R (c, filled area) and IL-22R1 (d, filled area) on keratinocyte single-cell suspensions compared with isotype control (dotted line) by flow cytometry. Double-label immunofluorescence shows that some CD11c+ dermal cells express IL-17R (e) but not IL-22R1 (f), while CD3+ cells in the dermis express neither receptor (g, h). Original magnification × 10.
We also noted expression of IL-17R and IL-22R1 on scattered dermal cells. To determine which dermal cells expressed these receptors, we performed double-label immunofluorescence: IL-17R was expressed by some CD11c+ dendritic cells (DCs) in the upper dermis (Fig. 2e) but not on CD3+ T cells (Fig. 2g) or CD163+ macrophages (data not shown). In contrast, IL-22R1 was not coexpressed on CD11c+ DCs (Fig. 2f), CD3+ T cells (Fig. 2h) or CD163+ macrophages (data not shown). However, vimentin+ fibroblasts in the dermis expressed IL-22R1 (data not shown).
Th1 and Th17 cytokines induce different keratinocyte gene signatures
To determine the effects of IL-17, IL-22 and IFN-γ on keratinocyte gene expression, we cultured primary human keratinocytes with IL-17 (200 ng mL−1), IL-22 (200 ng mL−1), IFN-γ (20 ng mL−1) or media alone for 24 h (n = 3). Interferons induce the transcription of large sets of known genes by activating STAT1 and ISGF3γ36 Hence, we assessed the response of keratinocytes to IFN-γ as both a positive control for the induction of expected genes, and to compare the gene sets that are regulated by Th1 vs. Th17 T-cell cytokines. We then performed microarray analysis filtering on genes that had a fold difference of > 1.5 compared with control with P < 0.05.
IFN-γ treatment induced keratinocyte expression of a large number of genes, with ~800 genes induced > 2-fold (Supplementary Table S1; see Supplementary material). The chemokines CXCL9, CXCL10 and CXCL11, all considered key IFN-γ-regulated chemokines, were induced > 7000 fold compared with control keratinocytes (Fig. 3). These three chemokines bind to CXCR3-bearing, activated T cells29 and are thought to be involved in T-cell trafficking to psoriatic dermis and overlying epidermis.37
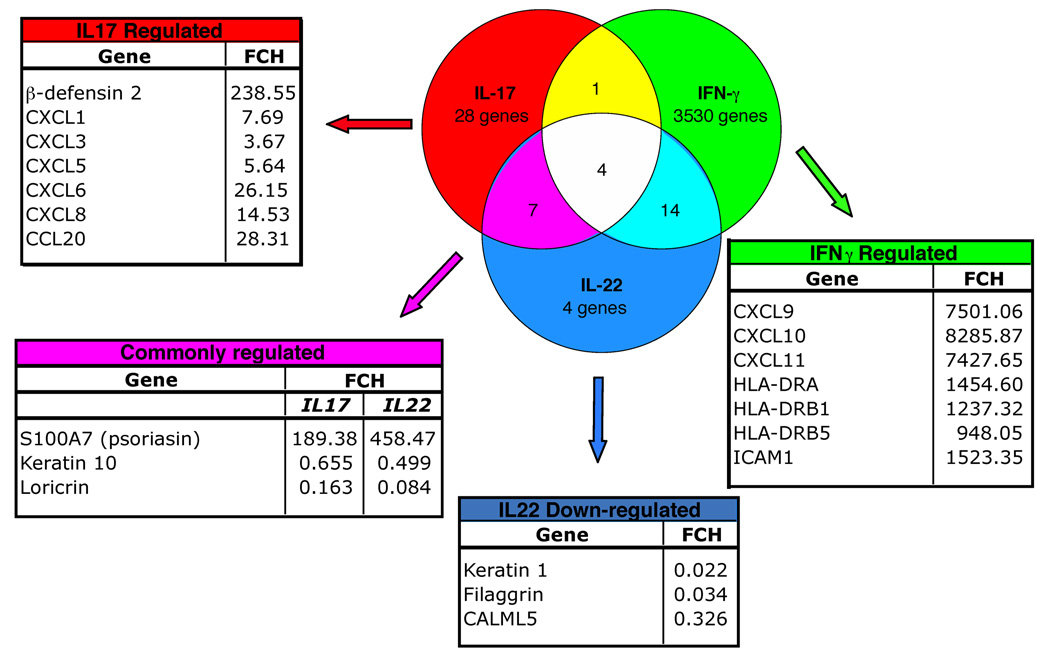
Fig 3.
Interleukin (IL)-17, IL-22 and interferon (IFN)-γ induce unique keratinocyte gene signatures. Venn diagram illustrates the number of keratinocyte genes regulated by IL-17, IL-22 or IFN-γ treatment. Surrounding tables list selected genes that are regulated by the cytokines with corresponding fold changes (FCH) from control gene expression levels.
We observed 28 keratinocyte genes that were regulated by IL-17 but not IL-22 nor IFN-γ, notably β-defensin 2 (DEFB4) and the neutrophil chemoattractants CXCL1, CXCL3, CXCL5, CXCL6 and CXCL8 (Fig. 3 and Supplementary Table S2; see Supplementary material). IL-17 also induced the expression of CCL20, a chemokine that preferentially attracts memory T cells and DCs,38,39 and is reportedly upregulated in psoriatic skin lesions.40
In contrast, the genes modulated by IL-22 but not IL-17 are part of the keratinocyte terminal differentiation pathway: keratin 1, filaggrin and CALML5 that were all downregulated by IL-22 (Fig. 3 and Supplementary Table S3; see Supplementary material).
Genes regulated by both IL-17 and IL-22 included the antimicrobial S100A7 (upregulated), and loricrin (downregulated) (Fig. 3).
Interleukin-17 upregulates the expression of immune cell chemoattractants that are increased in psoriatic lesions
To verify the microarray results, we quantified chemokine gene expression in treated keratinocytes by real-time RT-PCR, and normalized these expression values to the housekeeping gene HARP. We then assessed chemokine gene expression by treated keratinocytes as fold change difference from control keratinocyte values.
We confirmed that CXCL1, CXCL3, CXCL5, CXCL6 and CXCL8 were significantly induced by IL-17 treatment compared with IL-22 and IFN-γ (Fig. 4a). CCL20 gene expression was also found to be highly upregulated in IL-17-treated keratinocytes and was absent in cultures treated with either IL-22 or IFN-γ (Fig. 4a).
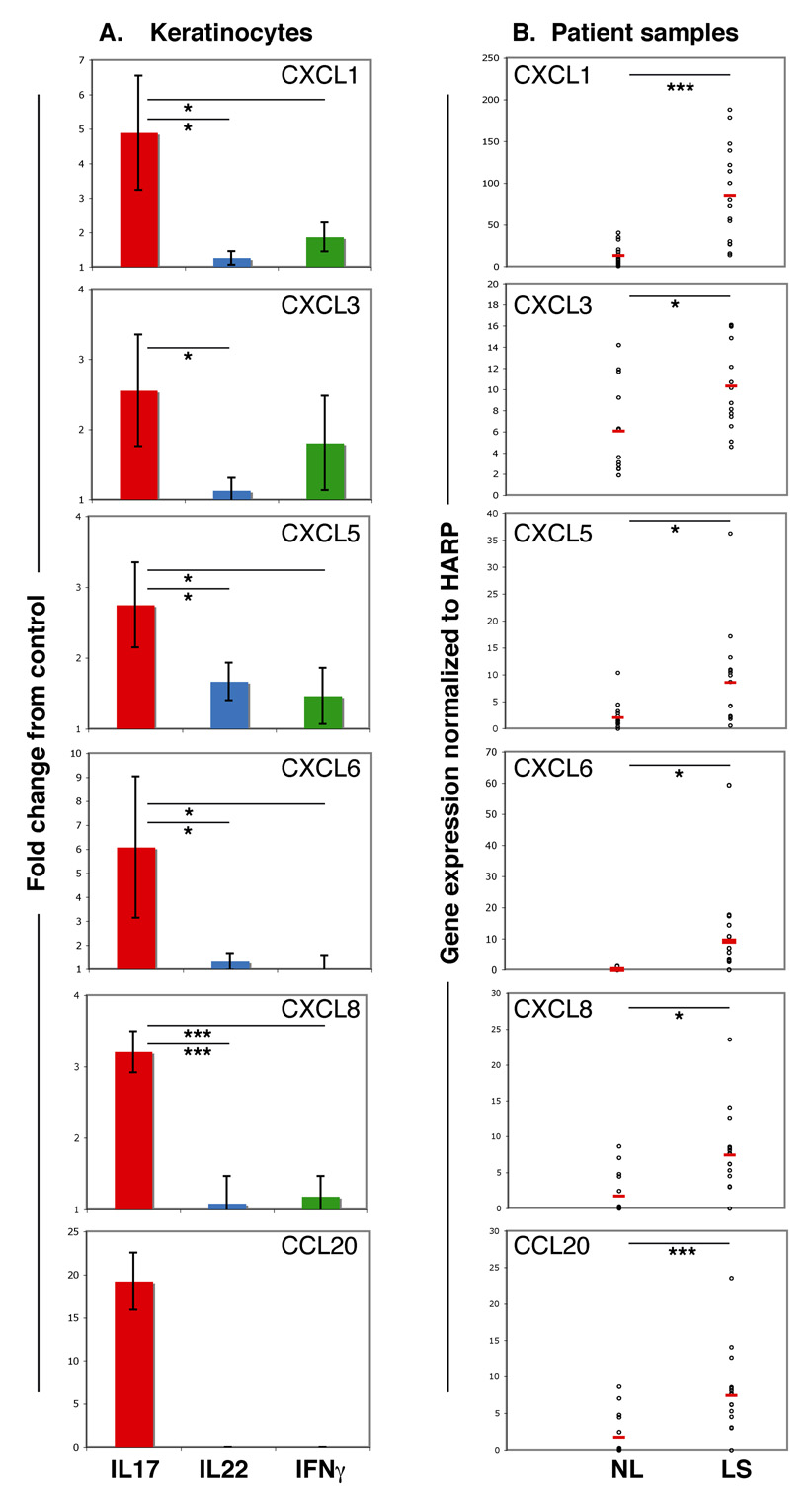
Fig 4.
Interleukin (IL)-17 induces the expression of immune chemoattractants that are upregulated in psoriatic skin. (a) Quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) analysis of chemokine gene expression in primary keratinocytes treated with IL-17, IL-22 or interferon (IFN)-γ expressed as fold change difference from control. Error bars indicate SD of biological triplicates. (b) RT-PCR quantification of chemokine gene expression normalized to HARP in paired samples of nonlesional (NL) and lesional skin (LS) from patients with psoriasis (n = 16). Open circles represent individual patients. Red lines represent the mean. Asterisks indicate statistical significance: *P < 0.05, **P < 0.01, ***P < 0.005.
To determine the extent to which these IL-17-induced chemokines were upregulated in psoriasis lesions, we performed quantitative RT-PCR on paired samples of lesional and nonlesional psoriatic skin (n = 16). We found that gene expression of all six chemokines was significantly increased in psoriatic lesions (Fig. 4b), paralleling results seen in IL-17-treated keratinocytes.
Neutrophils bearing CXCR1 and CXCR2 are present in psoriatic epidermis
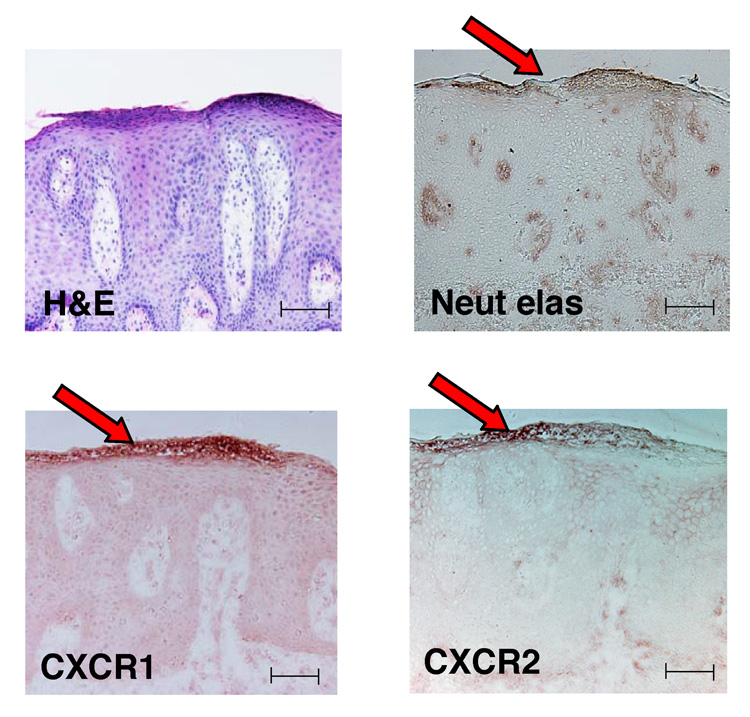
CXCL1, CXCL3, CXCL5, CXCL6 and CXCL8 all activate CXCR2 receptor, and CXCL8 can also activate CXCR1.30 We confirmed by immunohistochemistry that neutrophils found accumulating within subcorneal microabscesses bear both CXCR1 and CXCR2 receptors (Fig. 5). Our data suggest that these cells are present in psoriatic epidermis in response to ELR+ chemokines that are induced by IL-17.
Fig 5.
Neutrophils bearing CXCR1 and CXCR2 are present in psoriatic epidermis. Haematoxylin and eosin (H&E) staining and immunohistochemical analysis of psoriatic lesions (n = 5) demonstrate that neutrophils contained within subcorneal Munro’s microabscesses (red arrows) produce neutrophil elastase (Neut elas) and bear the chemokine receptors CXCR1 and CXCR2. Scale bars = 100 µm.
Interleukin-22 alters phenotype and gene expression of full-thickness skin equivalents
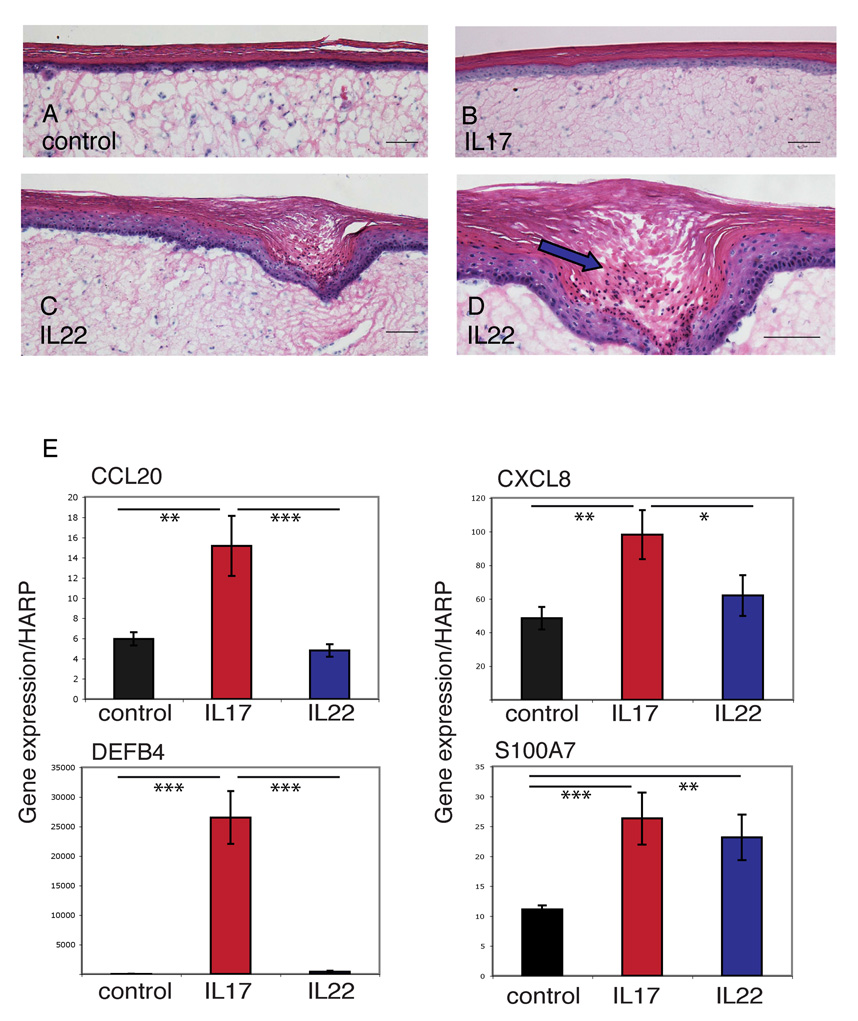
To determine the effect of IL-17 and IL-22 on human skin, we treated full-thickness skin equivalents with IL-17 (200 ng mL−1), IL-22 (200 ng mL−1), IFN-γ (20 ng mL−1) or control (n = 3). We found that the IL-22-treated skin equivalents developed acanthosis (Fig. 6c), downward epidermal projections (Fig. 6c) and parakeratotic areas (Fig. 6d) within 4 days, changes that were not observed with IL-17-treated (Fig. 6b) or control (Fig. 6a) equivalents. We then verified by quantitative RT-PCR that the genes we found regulated by IL-17 or IL-22 in cultured keratinocytes were similarly regulated in full-thickness skin equivalents. Indeed, as in cultured keratinocytes, IL-17 induced the expression of CCL20, β-defensin 2 (DEFB4) and CXCL8 in the skin equivalents, while both IL-17 and IL-22 upregulated S100A7 (Fig. 6e).
Fig 6.
Interleukin (IL)-22 induces acanthosis, parakeratosis and downward epidermal projections in full-thickness skin equivalents. Haematoxylin and eosin staining of full-thickness human skin equivalents treated without (a) or with (b–d) the indicated cytokines. Blue arrow (d) indicates parakeratosis. Scale bars = 100 µm. (E) Reverse transcriptase–polymerase chain reaction quantification of gene expression normalized to HARP by full-thickness skin equivalents. Error bars indicate SD of triplicate samples. Data shown are representative of three experiments. Asterisks indicate statistical significance: *P < 0.05, **P < 0.01, ***P < 0.005.
Discussion
Psoriasis is a complex skin disease, and a snapshot at any one time reveals the presence of a myriad of resident and inflammatory cells. The role of Th17 cells in inflammation is currently being investigated in several settings, including infection, inflammation and autoimmunity. IL-17, the cytokine product for which Th17 cells were named, is expressed at the genomic level in psoriasis,21 and Th17 cells have been identified from psoriasis lesions.18 IL-22, also an important product of Th17 cells, is upregulated in psoriasis lesions at the genomic level,18 and is implicated in keratinocyte hyperplasia.22,23 We were interested in evaluating the effects of these cytokines on normal keratinocytes, and determining whether the induced gene products were likewise upregulated in psoriasis lesions.
In this paper, we show data for the following potential inflammatory pathway: normal dermal CD4+ T cells produce IL-17 and IL-22, which bind to receptors present on normal keratinocytes, inducing key inflammatory products. IL-17 induces neutrophil, T-cell and DC chemokines, while IL-22 downregulates epidermal differentiation genes. Both these effects might be pathogenic during inflammation, contributing to the influx of neutrophils, memory T cells and DCs, and delaying keratinocyte differentiation, respectively. We also show that while IL-22 has been considered to be primarily a product of Th17 cells,15,16 in normal skin there appear to be three types of CD4+ T cells that can produce IL-22: Th17 cells, IFN-γ-producing Th1 cells, as well as a discrete CD4+ T-cell population – perhaps ‘Th22’ cells – that do not cosynthesize either IL-17 or IFN-γ.
Neutrophils are reportedly attracted to the skin by a number of chemotactic factors, including IL8/CXCL8 and gro/MGSA/CXCL1.41,42 We have shown that these, along with CXCL3, CXCL5 and CXCL6, are produced by keratinocytes in response to IL-17, are present in psoriatic lesions, and the receptors for these chemokines are on skin-infiltrating neutrophils. Neutrophils within the epidermis and stratum corneum (Kogoj and Munro’s microabscesses, respectively) produce reactive oxygen intermediates and proteolytic enzymes thought to promote local tissue destruction, unmask hidden antigens, or affect growth and differentiation of keratinocytes.43–46 Thus, keratinocytes in psoriatic lesions may release these neutrophil chemoattractants in response to T cell-derived inflammatory cytokines to drive neutrophil migration into the epidermis.47,48
IL-17 also induced CCL20 expression from keratinocytes. Although this has been previously reported (potentiated by tumour necrosis factor),40 here we show that while IL-17 is a major inducer of CCL20, IL-22 and IFN-γ are not. CCL20 binds to CCR6, which is expressed by highly differentiated resting memory T cells, B cells, DCs39 and Th17 cells themselves.14,49 This chemokine is upregulated in psoriasis, and CCL20-expressing keratinocytes have been shown to colocalize with CCR6-bearing T cells within the psoriatic epidermis.40 Thus, keratinocyte-derived CCL20 may play a central chemotactic role for inflammatory T cells and DCs, and may provide an autocrine loop to increase the influx of Th17 cells themselves.
IL-22 modulates a set of genes involved in keratinocyte mobility and terminal differentiation, and our data support the findings of other groups.50 While treatment of reconstituted human epidermis with IL-22 results in epidermal hyperplasia and hypogranulosis,22,23 we have extended these observations to show that treatment of full-thickness skin equivalents can induce parakeratosis, downward epidermal projections, and acanthosis within 4 days of treatment. Hence, it is very likely that IL-22 is responsible for disrupting normal keratinocyte differentiation in psoriasis.
There was little overlap between the lists of keratinocyte genes induced by these cytokines. This emphasizes the distinct roles that these cytokines, along with the T cells that produce them, play in disease pathogenesis. We confirm that IFN-γ induces keratinocyte expression of the CXCR3 ligands, CXCL9, CXCL10 and CXCL11. Each of these chemokines is upregulated in psoriatic lesions,37 and up to a third of T cells within psoriatic lesions express CXCR3.51
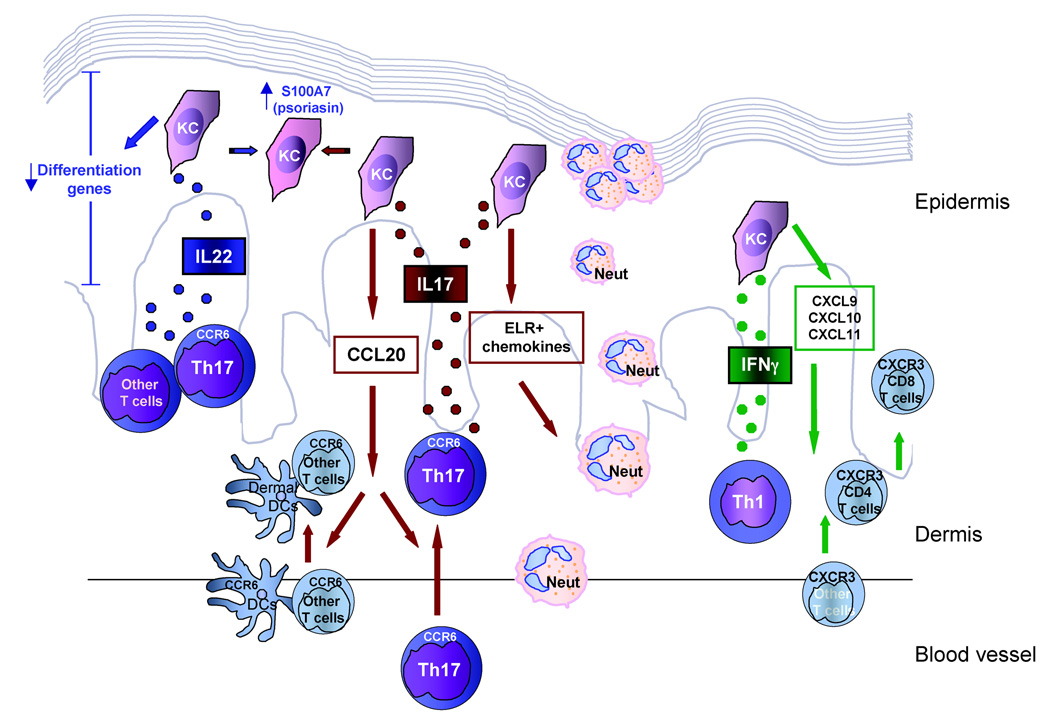
Figure 7 summarizes our working model of how these cytokines are likely to contribute to the disease manifestations observed in psoriasis vulgaris. Th17 cells produce both IL-17 and IL-22 cytokines. IL-17 is proinflammatory and drives the migration of neutrophils into the psoriatic lesion, contributing to the formation of Munro’s microabscesses. In addition, via CCL20, IL-17 brings CCR6-bearing DCs and memory T cells into the lesion, including additional Th17 cells. The other Th17 cytokine, IL-22, causes abnormal keratinocyte proliferation by downregulating genes that control terminal differentiation, resulting in the altered differentiation and parakeratosis. Both IL-17 and IL-22 can induce keratinocyte expression of the antimicrobial S100A7 (psoriasin). Lastly, the Th1-derived cytokine, IFN-γ, can regulate expression of cytokines that contribute to the trafficking of CXCR3+ T cells, including CD8+ T cells, into the psoriatic lesion.
Fig 7.
Working model of the likely contributions of the Th17 cytokines interleukin (IL)-17 and IL-22, and the Th1 cytokine interferon (IFN)-γ, to psoriasis. Th1, T helper 1 cells; Th17, T helper 17 cells; KC, keratinocyte; Neut, neutrophil; DC, dendritic cell. Th17 cells produce IL-17 that induces KCs to produce ELR+ chemokines and CCL20 bringing neutrophils, DCs and T cells into the psoriatic lesion. IL-22, produced by Th17 cells and other T cells, downregulates KC differentiation genes. IL-17 and IL-22 both upregulate S100A7 (psoriasin). Th1 cells release IFN-γ, inducing keratinocytes to produce CXCL9, CXCL10 and CXCL11 that cause influx of CXCR3-bearing T cells into the lesion.
Overall, genes that have altered expression in psoriasis cannot be explained by the effect of one cytokine on skin cells. While our data confirm the view that a number of genes upregulated in psoriasis lesions can be attributable to IFN-γ, we expand this concept in showing that other cytokines (IL-17 and IL-22) can also regulate other sets of psoriasis-related genes. We propose that the cytokines synthesized by Th1 and Th17 cells regulate distinct gene expression pathways in epidermal keratinocytes, and perhaps other skin-resident cells, and that the ‘transcriptome’ of psoriasis thus arises from the activation of multiple independent pathways.
Supplementary Material
1. Supplementary material.
The following supplementary material is available online for this article:
Table S1 Keratinocyte genes modulated by IFNγ treatment versus media control
Table S2 Keratinocyte genes modulated by IL-17 treatment versus control
Table S3 Keratinocyte genes modulated by IL-22 versus control
Acknowledgments
This publication was made possible by grant number 5UL1RR024143-02 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH. Information on NCRR is available at http://www.ncrr.nih.gov/. Information on Re-engineering the Clinical Research Enterprise can be obtained from http://nihroadmap.nih.gov/clinicalresearch/overview-translational.asp.
K.E.N. is supported by the Clinical Scholars Program at The Rockefeller University, L.C.Z. is supported by NIH MSTP grant GM07739, K.C.P. is supported by the Dana Foundation (Human Immunology Consortium Grant), and M.A.L. is supported by 1 K23 AR052404-01A1. We thank Patricia Gilleaudeau and Mary Whalen-Sullivan for the excellent care of our patients, A.N. LaBruna and D.M. Senderoff for their generous donation of abdominoplasty surgical waste, Yale Skin Diseases Research Center core facility for supplying primary keratinocytes, and Amgen, Inc. for providing us with monoclonal antihuman IL-17R antibody. The authors do not have any financial interest related to this work.
Footnotes
Conflicts of interest
None declared.
References
- 1.Ellis CN, Gorsulowsky DC, Hamilton TA, et al. Cyclosporine improves psoriasis in a double-blind study. JAMA. 1986;256:3110–3116. [PubMed] [Google Scholar]
- 2.Jegasothy BV, Ackerman CD, Todo S, et al. Tacrolimus (FK 506) – a new therapeutic agent for severe recalcitrant psoriasis. Arch Dermatol. 1992;128:781–785. [PMC free article] [PubMed] [Google Scholar]
- 3.Nicolas JF, Chamchick N, Thivolet J, et al. CD4 antibody treatment of severe psoriasis. Lancet. 1991;338:321. doi: 10.1016/0140-6736(91)90465-2. [DOI] [PubMed] [Google Scholar]
- 4.Gottlieb AB, Lebwohl M, Shirin S, et al. Anti-CD4 monoclonal antibody treatment of moderate to severe psoriasis vulgaris: results of a pilot, multicenter, multiple-dose, placebo-controlled study. J Am Acad Dermatol. 2000;43:595–604. doi: 10.1067/mjd.2000.107945. [DOI] [PubMed] [Google Scholar]
- 5.Barker JN, Karabin GD, Stoof TJ, et al. Detection of interferon-gamma mRNA in psoriatic epidermis by polymerase chain reaction. J Dermatol Sci. 1991;2:106–111. doi: 10.1016/0923-1811(91)90019-t. [DOI] [PubMed] [Google Scholar]
- 6.Szabo SK, Hammerberg C, Yoshida Y, et al. Identification and quantitation of interferon-gamma producing T cells in psoriatic lesions: localization to both CD4+ and CD8+ subsets. J Invest Dermatol. 1998;111:1072–1078. doi: 10.1046/j.1523-1747.1998.00419.x. [DOI] [PubMed] [Google Scholar]
- 7.Uyemura K, Yamamura M, Fivenson DF, et al. The cytokine network in lesional and lesion-free psoriatic skin is characterized by a T-helper type 1 cell-mediated response. J Invest Dermatol. 1993;101:701–705. doi: 10.1111/1523-1747.ep12371679. [DOI] [PubMed] [Google Scholar]
- 8.Harrington LE, Hatton RD, Mangan PR, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 9.Harrington LE, Mangan PR, Weaver CT. Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr Opin Immunol. 2006;18:349–356. doi: 10.1016/j.coi.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Langrish CL, McKenzie B, et al. Anti-IL-23 therapy inhibits multiple inflammatory pathways and ameliorates autoimmune encephalomyelitis. J Clin Invest. 2006;116:1317–1326. doi: 10.1172/JCI25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettelli E, Carrier Y, Gao W, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Tato CM, Muul L, et al. Distinct regulation of interleukin-17 in human T helper lymphocytes. Arthritis Rheum. 2007;56:2936–2946. doi: 10.1002/art.22866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aggarwal S, Ghilardi N, Xie MH, et al. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 14.Annunziato F, Cosmi L, Santarlasci V, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang SC, Tan XY, Luxenberg DP, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y, Danilenko DM, Valdez P, et al. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 17.Lee E, Trepicchio WL, Oestreicher JL, et al. Increased expression of interleukin 23 p19 and p40 in lesional skin of patients with psoriasis vulgaris. J Exp Med. 2004;199:125–130. doi: 10.1084/jem.20030451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowes MA, Kikuchi T, Fuentes-Duculan J, et al. Psoriasis vulgaris lesions contain discrete populations of Th1 and Th17 T cells. J Invest Dermatol. 2008;128:1207–1211. doi: 10.1038/sj.jid.5701213. [DOI] [PubMed] [Google Scholar]
- 19.Zaba LC, Cardinale I, Gilleaudeau P, et al. Amelioration of epidermal hyperplasia by TNF inhibition is associated with reduced Th17 responses. J Exp Med. 2007;204:3183–3194. doi: 10.1084/jem.20071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boniface K, Guignouard E, Pedretti N, et al. A role for T cell-derived interleukin 22 in psoriatic skin inflammation. Clin Exp Immunol. 2007;150:407–415. doi: 10.1111/j.1365-2249.2007.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Chen X, Liu Z, et al. Expression of Th17 cytokines in skin lesions of patients with psoriasis. J Huazhong Univ Sci Technolog Med Sci. 2007;27:330–332. doi: 10.1007/s11596-007-0329-1. [DOI] [PubMed] [Google Scholar]
- 22.Boniface K, Bernard FX, Garcia M, et al. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J Immunol. 2005;174:3695–3702. doi: 10.4049/jimmunol.174.6.3695. [DOI] [PubMed] [Google Scholar]
- 23.Sa SM, Valdez PA, Wu J, et al. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J Immunol. 2007;178:2229–2240. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- 24.Ma HL, Liang S, Li J, et al. IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation. J Clin Invest. 2008;118:597–607. doi: 10.1172/JCI33263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson NJ, Boniface K, Chan JR, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8:950–957. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 26.Linden A, Hoshino H, Laan M. Airway neutrophils and interleukin-17. Eur Respir J. 2000;15:973–977. doi: 10.1034/j.1399-3003.2000.15e28.x. [DOI] [PubMed] [Google Scholar]
- 27.McAllister F, Henry A, Kreindler JL, et al. Role of IL-17A, IL-17F, and the IL-17 receptor in regulating growth-related oncogene-alpha and granulocyte colony-stimulating factor in bronchial epithelium: implications for airway inflammation in cystic fibrosis. J Immunol. 2005;175:404–412. doi: 10.4049/jimmunol.175.1.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laan M, Cui ZH, Hoshino H, et al. Neutrophil recruitment by human IL-17 via C-X-C chemokine release in the airways. J Immunol. 1999;162:2347–2352. [PubMed] [Google Scholar]
- 29.Colobran R, Pujol-Borrell R, Armengol MP, et al. The chemokine network. I. How the genomic organization of chemokines contains clues for deciphering their functional complexity. Clin Exp Immunol. 2007;148:208–217. doi: 10.1111/j.1365-2249.2007.03344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weathington NM, Blalock JE. The biology of CXC chemokines and their receptors. In: Schwiebert LM, editor. Chemokines, Chemokine Receptors and Disease (Current Topics in Membranes Vol. 55) New York: Academic Press; 2005. pp. 49–71. [Google Scholar]
- 31.Fossiez F, Djossou O, Chomarat P, et al. T cell interleukin-17 induces stromal cells to produce proinflammatory and hematopoietic cytokines. J Exp Med. 1996;183:2593–2603. doi: 10.1084/jem.183.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albanesi C, Cavani A, Girolomoni G. IL-17 is produced by nickel-specific T lymphocytes and regulates ICAM-1 expression and chemokine production in human keratinocytes: synergistic or antagonist effects with IFN-gamma and TNF-alpha. J Immunol. 1999;162:494–502. [PubMed] [Google Scholar]
- 33.Chamian F, Lowes MA, Lin SL, et al. Alefacept reduces infiltrating T cells, activated dendritic cells, and inflammatory genes in psoriasis vulgaris. Proc Natl Acad Sci USA. 2005;102:2075–2080. doi: 10.1073/pnas.0409569102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X, Krueger JG, Kao MC, et al. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics. 2003;13:69–78. doi: 10.1152/physiolgenomics.00157.2002. [DOI] [PubMed] [Google Scholar]
- 35.Suarez-Farinas M, Pellegrino M, Wittkowski KM, et al. Harshlight: a ‘corrective make-up’ program for microarray chips. BMC Bioinformatics. 2005;6:294. doi: 10.1186/1471-2105-6-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baechler EC, Batliwalla FM, Karypis G, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rottman JB, Smith TL, Ganley KG, et al. Potential role of the chemokine receptors CXCR3, CCR4, and the integrin alphaEbeta7 in the pathogenesis of psoriasis vulgaris. Lab Invest. 2001;81:335–347. doi: 10.1038/labinvest.3780242. [DOI] [PubMed] [Google Scholar]
- 38.Dieu MC, Vanbervliet B, Vicari A, et al. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J Exp Med. 1998;188:373–386. doi: 10.1084/jem.188.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao F, Rabin RL, Smith CS, et al. CC-chemokine receptor 6 is expressed on diverse memory subsets of T cells and determines responsiveness to macrophage inflammatory protein 3 alpha. J Immunol. 1999;162:186–194. [PubMed] [Google Scholar]
- 40.Homey B, Dieu-Nosjean MC, Wiesenborn A, et al. Up-regulation of macrophage inflammatory protein-3 alpha/CCL20 and CC chemokine receptor 6 in psoriasis. J Immunol. 2000;164:6621–6632. doi: 10.4049/jimmunol.164.12.6621. [DOI] [PubMed] [Google Scholar]
- 41.Schroder JM, Gregory H, Young J, et al. Neutrophil-activating proteins in psoriasis. J Invest Dermatol. 1992;98:241–247. doi: 10.1111/1523-1747.ep12556058. [DOI] [PubMed] [Google Scholar]
- 42.van Damme J, van Beeumen J, Opdenakker G, et al. A novel, NH2-terminal sequence-characterized human monokine possessing neutrophil chemotactic, skin-reactive, and granulocytosis-promoting activity. J Exp Med. 1988;167:1364–1376. doi: 10.1084/jem.167.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer-Hoffert U, Wingertszahn J, Wiedow O. Human leukocyte elastase induces keratinocyte proliferation by epidermal growth factor receptor activation. J Invest Dermatol. 2004;123:338–345. doi: 10.1111/j.0022-202X.2004.23202.x. [DOI] [PubMed] [Google Scholar]
- 44.Glinski W, Barszcz D, Janczura E, et al. Neutral proteinases and other neutrophil enzymes in psoriasis, and their relation to disease activity. Br J Dermatol. 1984;111:147–154. doi: 10.1111/j.1365-2133.1984.tb04037.x. [DOI] [PubMed] [Google Scholar]
- 45.Ludolph-Hauser D, Schubert C, Wiedow O. Structural changes of human epidermis induced by human leukocyte-derived proteases. Exp Dermatol. 1999;8:46–52. doi: 10.1111/j.1600-0625.1999.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 46.Rogalski C, Meyer-Hoffert U, Proksch E, et al. Human leukocyte elastase induces keratinocyte proliferation in vitro and in vivo. J Invest Dermatol. 2002;118:49–54. doi: 10.1046/j.0022-202x.2001.01650.x. [DOI] [PubMed] [Google Scholar]
- 47.Barker JN, Jones ML, Mitra RS, et al. Modulation of keratinocyte-derived interleukin-8 which is chemotactic for neutrophils and T lymphocytes. Am J Pathol. 1991;139:869–876. [PMC free article] [PubMed] [Google Scholar]
- 48.Terui T, Ozawa M, Tagami H. Role of neutrophils in induction of acute inflammation in T-cell-mediated immune dermatosis, psoriasis: a neutrophil-associated inflammation-boosting loop. Exp Dermatol. 2000;9:1–10. doi: 10.1034/j.1600-0625.2000.009001001.x. [DOI] [PubMed] [Google Scholar]
- 49.Singh SP, Zhang HH, Foley JF, et al. Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J Immunol. 2008;180:214–221. doi: 10.4049/jimmunol.180.1.214. [DOI] [PubMed] [Google Scholar]
- 50.Wolk K, Witte E, Wallace E, et al. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur J Immunol. 2006;36:1309–1323. [Google Scholar]
- 51.Teraki Y, Miyake A, Takebayashi R, et al. Homing receptor and chemokine receptor on intraepidermal T cells in psoriasis vulgaris. Clin Exp Dermatol. 2004;29:658–663. doi: 10.1111/j.1365-2230.2004.01638.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. Supplementary material.
The following supplementary material is available online for this article:
Table S1 Keratinocyte genes modulated by IFNγ treatment versus media control
Table S2 Keratinocyte genes modulated by IL-17 treatment versus control
Table S3 Keratinocyte genes modulated by IL-22 versus control