AIM2 recognizes cytosolic dsDNA and forms a caspase-1 activating inflammasome with ASC (original) (raw)
. Author manuscript; available in PMC: 2009 Sep 26.
Published in final edited form as: Nature. 2009 Jan 21;458(7237):514–518. doi: 10.1038/nature07725
Abstract
The innate immune system senses nucleic acids via germ-line encoded pattern recognition receptors. RNA is sensed via Toll-like receptor (TLR)−3, −7 and −8 or by the RNA helicases RIG-I and MDA-51. Little is known about sensors for cytoplasmic DNA which trigger antiviral and/or inflammatory responses2–6. The best characterized of these responses involves activation of the TANK-binding kinase (TBK1)-Interferon Regulatory Factor (IRF)-3 signaling axis to trigger transcriptional induction of IFN〈/® genes2,3. A second, less well-defined pathway leads to the activation of an ‘inflammasome’ which via caspase-1, controls the catalytic cleavage of the pro-forms of the cytokines IL-1β and IL-186,7. Here we identify the IFI20X/IFI16 (PYHIN) family member8, absent in melanoma 2 (AIM2), as a receptor for cytosolic DNA which regulates caspase-1. The HIN200 domain of AIM2 binds to DNA, while the PYD domain (but not that of the other PYHIN family members) associates with the adapter molecule ASC to activate both NF-κB and caspase-1. Knockdown of AIM2 abrogates caspase-1 activation in response to cytoplasmic dsDNA and the dsDNA virus, vaccinia. Collectively, these observations identify AIM2 as a novel receptor for cytoplasmic DNA, which forms an inflammasome with the ligand and ASC to activate caspase-1.
Our current understanding of the mechanisms sensing cytoplasmic DNA is limited9. A candidate receptor called DAI (DNA-dependent Activator of IFN-regulatory factors) has been implicated in the DNA-induced type I IFN pathway4. The NLR family member, NLRP3 has also been shown to activate caspase-1 in response to internalized adenoviral DNA6. Caspase-1 activation in response to transfected bacterial, viral, mammalian or synthetic DNA however, does not involve NLRP3, although the adapter molecule ASC is required6,7.
We hypothesized that an upstream activator of this dsDNA activated ASC pathway would contain a pyrin domain (PYD) for homotypic interaction with ASC and at least one additional domain, for direct binding to DNA or for association with an upstream receptor. In addition to NLRP310, NLRP611 and NLRP1212 have previously been shown to associate with ASC. Although ASC-deficient macrophages failed to activate caspase-1 and trigger IL-1β release in response to poly(dA-dT) · poly(dA-dT) [hereafter referred to as poly(dA-dT)])2, macrophages lacking NLRP3, −6 and −12 responded normally (Fig. 1a). Surprisingly, we found that macrophages lacking ASC had higher levels of IFNβ and IL-6 in response to poly(dA-dT), which was not observed in cells lacking NLRP3, the IL1R or to a lesser extent caspase-1 (Supplementary Fig. 1a–d). Poly(dA-dT) induced cell death also occurred in an ASC-dependent manner (Supplementary Fig. 1e–f). We speculate, that the increased cytokine production in ASC-deficient cells relates to their resistance to poly(dA-dT) induced cell death. In addition to poly(dA-dT), dsDNA from natural sources activated caspase-1 cleavage (Supplementary Fig. 2a–b). In contrast, a small immunostimulatory oligonucleotide (ISD)3, long ssDNA (poly(dI)), transfected dsRNA or the ssRNA virus Sendai virus failed to trigger this response in NLRP3-deficient macrophages (Supplementary Fig. 2c).
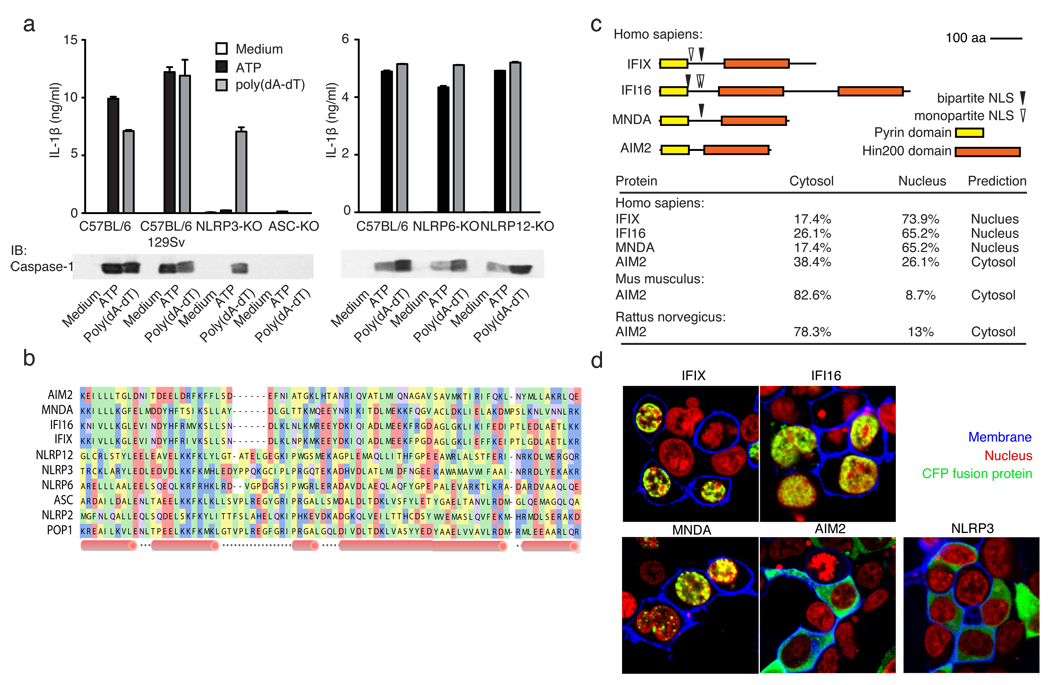
Figure 1. poly(dA-dT)-induced inflammasome activation.
a, LPS-primed macrophages from wild type or inflammasome deficient mice were stimulated as indicated and supernatants examined for IL-1β by ELISA or for cleaved caspase-1 by immunoblot. b, A multiple sequence alignment of human PYHIN and select NLR PYD domains, c, Domain structures of human PYHINs, with predicted nuclear localization signals and subcellular localizations (lower panel). d, Subcellular localization of CFP-tagged IFIX, IFI16, MNDA, AIM2 or NLRP3 (green) in 293T cells. Fluorescent choleratoxin stained membranes (blue) and DRAQ5-stained nuclei (red). Data from one experiment of three is shown (a, d).
Searching the PFAM database13 we identified several PYD-domain containing proteins, which also contained a HIN200 domain, previously shown to bind DNA14,. In humans, the HIN200 family consists of four members15; IFIX16, IFI1617, MNDA18 and AIM219. A multiple sequence alignment of PYD domains from these proteins with PYD domains from some of the NLRs is shown (Fig. 1b). Sequence analysis of IFIX, IFI16 and MNDA predicted their nuclear localization, in contrast to AIM2, which was predicted to be cytosolic (Fig. 1c, lower panel). Consistent with these predictions, fluorescent protein chimeras of IFIX, IFI16 and MNDA localized to the nucleus, while AIM2 was almost exclusively cytoplasmic (Fig. 1d).
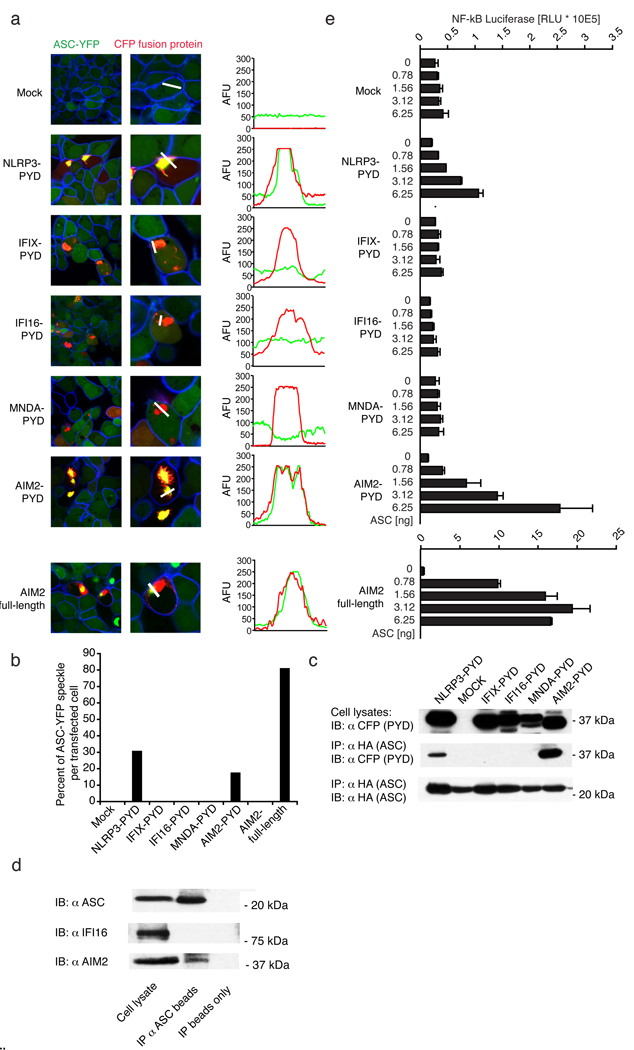
To study the possibility that these PYHIN proteins associated with ASC, we generated C-terminally tagged CFP PYD-domain fusions (which lacked the putative nuclear localization sequences identified above). Indeed, all of the PYD-CFP fusions were localized in the cytoplasm (Supplementary Fig. 3). To test whether induced clustering of the PYD-CFP fusions leads to association with ASC-YFP, we utilized a HEK293 cell line that stably expressed ASC-YFP at low enough levels to be polydispersed throughout the cytoplasm (Fig. 2a, Mock). Indeed, overexpression of the NLRP3-CFP-tagged PYD domain led to the formation of large cytosolic aggregates, which co-aggregated ASC-YFP (Fig. 2a). Notably, in most transfected cells, extensive intracellular co-localization with ASC-YFP was observed with a complete loss of cytoplasmic distribution of ASC-YFP10,11,20. Of all the PYHIN-PYD proteins tested, only AIM2-PYD led to complex formation with ASC (Fig. 2a, b and Supplementary Fig. 4). Similar results were obtained with full-length AIM2-CFP but not full-length IFIX, IFI16 or MNDA, which were all localized to the nucleus (Fig. 2a, b and Supplementary Fig. 5a, b). Additionally, only AIM2-PYD and NLRP3-PYD were found to bind HA-tagged ASC in co-immunoprecipitation studies (Fig. 2c). Finally, endogenous ASC associated with endogenous AIM2, but not IFI16 in primed THP-1 cells (Fig. 2d).
Figure 2. AIM2 interacts with ASC.
a, ASC-YFP-expressing cells (green) were transfected as indicated and imaged by confocal microscopy. Fluorescence intensities of green (YFP-ASC) and red (PYD-CFP) channels were quantified along white lines and b, the percentage of ASC-YFP speckles shown. c, 293T cells transfected with HA-ASC and CFP-tagged constructs as in (a) were immunoprecipitated (IP) with anti-HA and immunoblotted (WB) as indicated (lower panel). d, ASC was immunoprecipitated from Sendai virus primed THP-1 cells and ASC, IFI16 and AIM2 examined by immunoblotting. The band above the AIM2 band in the ASC immunoprecipitation corresponds to the heavy chain of the anti-ASC antibody e, NF-κB luciferase reporter gene activity was measured upon transfection with the indicated plasmids. Data of one representative experiment out of three (a, b, c and d) or two (e) are depicted.
To examine the functional relevance of AIM2-ASC complex formation, we examined NF-κB reporter gene activity in cells overexpressing the PYD-PYHIN proteins in the presence of ASC. Only NLRP3-PYD and AIM2-PYD led to potent NF-κB activation (Fig. 2e). The effect of full length AIM2 was even more dramatic (Fig. 2e, bottom panel). The full-length versions of IFIX, IFI16 and MNDA failed to activate NF-κB (Supplementary Fig. 5c). ASC was absolutely required, since no substantial NF-κB reporter activity was observed in cells not transfected with ASC. No substantial activation of the IFNβ promoter reporter gene was observed with any of the PYHIN family members (Supplementary Fig. 6).
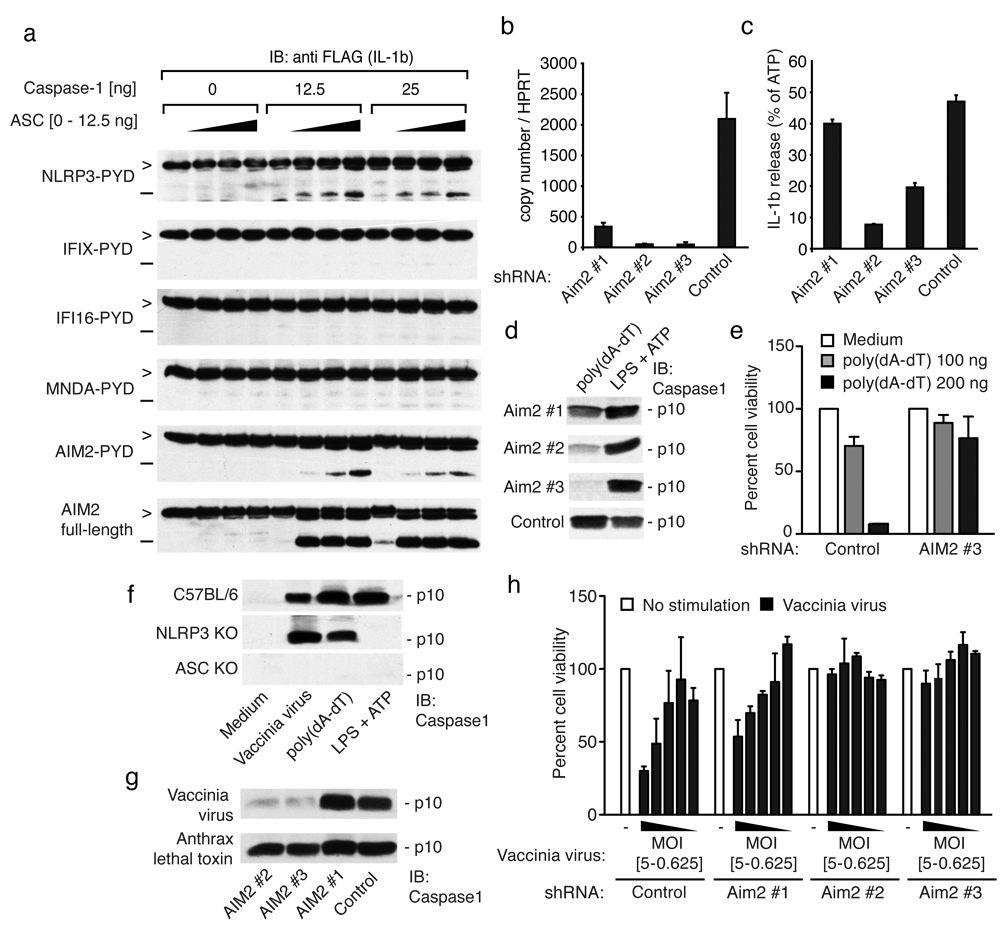
We next examined whether the AIM2-ASC complex could lead to the formation of a functional inflammasome complex and caspase-1-dependent maturation of pro-IL-1β. We employed a transient transfection assay overexpressing the respective proteins of interest in the presence of ASC, caspase-1 and flag-tagged pro-IL-1β in 293T cells and monitored the cleavage of pro-IL-1β by immunoblotting. Among the PYD proteins tested, only that of NLRP3-PYD and AIM2-PYD induced maturation of pro-IL-1β, when ASC and caspase-1 were co-expressed (Fig. 3a). Full-length AIM2 was even more potent than AIM2-PYD (Fig. 3a, lower panel). Neither the PYD domain nor the full-length versions of IFIX, IFI16 or MNDA induced IL-1β cleavage (Supplementary Fig. 7).
Figure 3. AIM2 is required for poly(dA-dT)- and vaccinia virus-triggered inflammasome activation.
a, 293T cells were transfected as indicated and cell lysates immunoblotted for pro-IL-1β (>) and cleaved IL-1β (−). b, B6-MCLs were transduced with lentiviral vectors encoding shRNAs as indicated and AIM2/HPRT1 measured by QPCR. c, LPS-primed cells as in (b) were stimulated as indicated for 6 hrs and IL-1β in supernatants measured by ELISA. The poly(dA-dT) triggered IL-1β release was normalized to the ATP induced IL-1β levels. Absolute values for ATP-triggered IL-1β release were 1790, 2078, 2676 and 1119 pg/ml (AIM2#1, AIM2#2, AIM2#3 and Control) respectively. d, Macrophages as in (b) were treated as indicated and cleavage of caspase-1 measured by immunoblotting after 6h. e, Macrophages transduced with shRNA were transfected as indicated and cells counted 24h later. f, Macrophages from the indicated strains were treated as indicated and cleavage of caspase-1 measured after 6h. g, NLRP3-deficient shRNA expressing macrophages as in (b) were infected as indicated and assessed for cleavage of caspase-1 after 6h. h, Cells as in (b) were infected as indicated and cell survival measured by calcein AM staining 24 h later. One representative experiment out of three (a, d, e, f, g and h) four (c) or five (b) is depicted.
To study the role of AIM2 in cells with a functional poly(dA-dT)-triggered or dsDNA virus induced inflammasome complex, we used lentiviruses encoding shRNAs to knock down AIM2 in immortalized murine macrophage cell lines (B6-MCLs or N3-KO-MCLs)7. AIM2 was expressed constitutively in both primary macrophages and in B6-MCLs and was further induced by poly(dA-dT) or Sendai virus (Supplementary Fig. 8). Three different shRNAs were tested, of which two (shRNA AIM2 #2 and AIM2 #3) resulted in a strong reduction of AIM2 expression (Fig. 3b). Knocking down AIM2, but not an unrelated gene, resulted in a strong attenuation of poly(dA-dT)-mediated IL-1β release (Fig. 3c) and caspase-1 cleavage (Fig. 3d). Targeting AIM2 in THP1 cells using siRNA corroborated these findings (Supplementary Fig. 8d and e). Moreover and consistent with what we had seen in ASC-deficient macrophages (Supplementary Fig. 1), knocking down AIM2 resulted in a marked enhancement of poly(dA-dT)-mediated type I IFN induction (Supplementary Fig. 8b). This effect was specific since the IFNβ response to Sendai virus was unaffected (Supplementary Fig. 8c). In addition, and in agreement with the results obtained in ASC-deficient macrophages, macrophages that were targeted with AIM2 shRNAs were resistant to poly(dA-dT) triggered cell death (Fig. 3e). We also examined the role of AIM2 in the recognition of the dsDNA virus vaccinia. Similar to what we had observed with transfected poly(dA-dT), vaccinia virus-induced caspase-1 cleavage occurred in an ASC-dependent but NLRP3-independent manner (Fig. 3f). This effect was also dependent on AIM2, since shRNA-mediated knock down of AIM2 impaired vaccinia virus induced caspase-1 cleavage but not that induced by anthrax lethal toxin (Fig. 3g). Moreover, knock down of a control protein did not affect caspase-1 cleavage after vaccinia virus infection. Vaccinia virus-triggered cell death was also strongly reduced in AIM2 shRNA targeted macrophages, but not in control macrophages (Fig. 3h). Altogether, these results indicated that AIM2 controlled inflammasome activation and cell death in response to dsDNA and the dsDNA virus vaccinia.
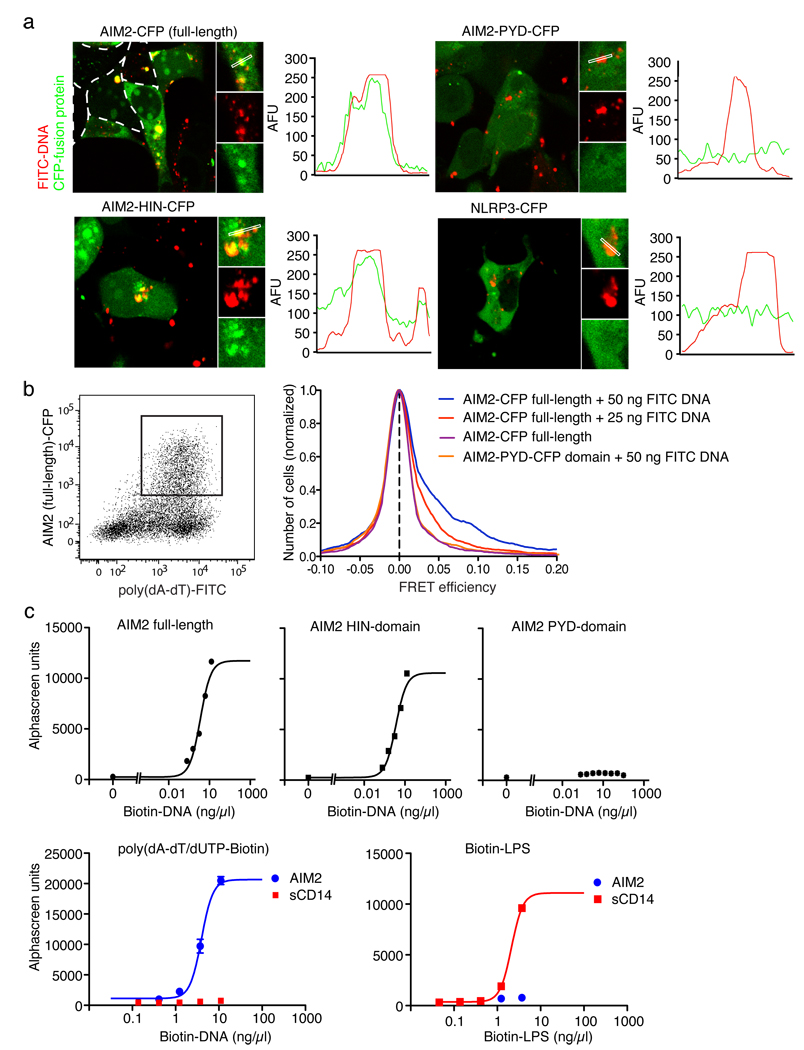
To examine if AIM2 could be involved in the recognition of dsDNA directly, we generated fluorescein-labeled poly(dA-dT), (FITC-dsDNA) and co-transfected FITC-dsDNA together with CFP-tagged versions of full-length AIM2, AIM2-HIN domain, AIM2-PYD domain or full-length NLRP3. While cells expressing NLRP3 or AIM2-PYD showed no co-localization of the respective proteins with FITC-dsDNA, full-length AIM2 and AIM2-HIN domain showed extensive co-localization with FITC-dsDNA and led to the formation of DNA/protein aggregates in the cytosol (Fig. 4a). We used single cell flow cytometry fluorescence resonance energy transfer (FRET) measurements to quantify these interactions (Fig. 4b). A dose-dependent increase in FRET between full-length AIM2 and FITC-dsDNA was seen, while AIM2-PYD did not lead to measurable FRET. Other proteins such as NLRP3 or IFI16 did not show any FRET (data not shown). Additionally, binding studies using purified AIM2, AIM2-HIN domain and AIM2-PYD domain with biotinylated poly(dA-dT) (biotin-dsDNA) revealed that AIM2 directly interacted with poly(dA-dT) with high affinity; only full-length AIM2 or the AIM2-HIN domain were able to bind biotin-dsDNA (Fig. 4c). Binding of poly(dA-dT) to AIM2 was specific, since AIM2 did not bind biotin-LPS, which bound to soluble CD14 under similar assay conditions (Fig. 4c).
Figure 4. AIM2 binds dsDNA via its HIN domain.
a, 293T cells were transfected with CFP-tagged AIM2 or NLRP3 (red) as indicated together with fluorescein labeled poly(dA-dT) (FITC-DNA, green) and imaged by confocal microscopy. Fluorescence intensities of the green (FITC-DNA) and red (CFP fusion protein) channels were quantified along selected lines. b, 293T cells were transfected as indicated with FITC-DNA or unlabeled DNA. Cells were analyzed by flow cytometry after 24 h and CFP and FITC positive cells were gated (left panel) and analyzed for FRET efficiency on a cell-by-cell basis. Calculated FRET efficiency histograms are shown (right panel). c, AlphaScreen assessment of AIM2 full length, AIM2-HIN domain or AIM2-PYD domain binding to poly(dA-dT)/Biotin-dUTP (Biotin-DNA) (upper panel) and AIM2 or sCD14 binding to Biotin-DNA or biotin LPS (lower panel). Representative data from 2 (d) or three (a, b, c) independent experiments are shown.
Collectively, these data identify AIM2 as a receptor for cytosolic dsDNA, which forms a novel inflammasome complex with ASC to activate caspase-1-mediated processing of IL-1β. Our data also indicate that the activation of the AIM2 inflammasome is important in innate immunity to vaccinia virus. Since bacterial pathogens such as Francisella tularensis 21 and aberrant host DNA in pathological autoimmunity22 also trigger the IL1β pathway, it will also be important to define the role of AIM2 in these responses. Further characterization of the AIM2 inflammasome as a sensor of microbial, as well as host DNA therefore, may enable the rational design of new therapies and treatments for infectious as well as autoimmune diseases.
Methods Summary
Reagents
All cDNAs were cloned by PCR from cDNA into pEFBOS-C-term-CFP and subcloned into pEFBOS-C-term-FLAG/HIS. Biotinylated and FITC-labeled poly(dA-dT) were made by adding Biotin-dUTP or FITC-dUTP (Fermentas) in a molar ratio of 1:8 to dTTP in the enzymatic synthesis of poly(dA-dT) as described23. Vaccinia virus (Western Reserve strain) was from K. Rock (UMASS, Worcester, MA). Anti-human AIM2 antibody (3B10) was from R. Johnstone (The Peter MacCallum Cancer Institute, Victoria, Australia).
Mice
NLRP3 −/− and Pycard −/− mice were as previously described24. Both strains, as well as NLRP6−/− and NLRP12−/− mice were from Millennium Pharmaceuticals (Cambridge, MA). Caspase-1-deficient mice were from R. Flavell (Yale University, New Haven, CT). IL-1R-deficient (Il1r1 −/−) were from Jackson Laboratories (Bar Harbor, ME).
Cell culture and stimulation
Bone marrow derived macrophages were stimulated as indicated. Poly(dA-dT) DNA and all other DNAs were transfected using Lipofectamine 2000 at a concentration of 1 µg/ml.
ELISA
Cell culture supernatants were assayed for IL-1β using ELISA kits from BD Biosciences (Franklin Lakes, NJ).
Confocal microscopy
Confocal microscopy was performed on a Leica SP2 AOBS confocal laser scanning microscope.
Flow cytometry Fluorescence Resonance Energy Transfer (FRET)
FRET efficiencies were calculated on a cell-by-cell basis25 and histograms were plotted with GraphPad Prism 5.01 (GraphPad Software).
Immunoblot analysis
Immunoblot analysis was conducted as previously described7.
Quantitative real-time PCR
Quantitative RT-PCR analysis performed as previously described26.
shRNA mediated silencing
Lentiviral shRNAs targeting AIM2 were obtained from OpenBiosystems and shRNA silencing carried out as described (http://www.broad.mit.edu/genome_bio/trc/publicProtocols.html).
AlphaScreen Assay
The AlphaScreen (amplified luminescent-proximity homogeneous assay) was set up as an association assay and read with the Envision HT microplate reader (Perkin Elmer).
Reporter assays
Reporter assays for NF-κB or IFN® luciferase reporters were carried out as previously described26.
ONLINE METHODS
Plasmid constructs
Full length human AIM2 (1–356; based on BC010940.1), AIM2-PYD (1–83), AIM2-HIN (148–356), IFIX (1–461; based on NM_198929), IFIX-PYD (1–84), IFI16 (1–729; based on NM_005531), IFI16-PYD (1–84), MNDA (1–407), MNDA-PYD (1–84; based on NM_002432) and NLRP3-PYD (1–87; based on NM_004895.3) were cloned by PCR from cDNA into pEFBOS-C-term-CFP using PCR-generated XhoI and BamHI or BglII restriction sites. AIM2 full length, AIM2-PYD and AIM2-HIN were subcloned into pEFBOS-C-term-FLAG/HIS using XhoI and BamHI. Murine pro-IL-1β (1–269) was obtained by PCR from cDNA and fused into pEFBOS-C-term-GLuc/FLAG using XhoI and BglII/BamHI. Expression plasmids (pCI) encoding human Caspase-1 and ASC-HA were from Millenium Pharmaceuticals (Cambridge, MA).
Reagents
ATP, LPS, poly(dA-dT), were from Sigma-Aldrich. A555-conjugated choleratoxin B was obtained from Molecular Probes, Invitrogen. DRAQ5 was from Biostatus. Biotinylated and FITC-labeled poly(dA-dT) were made by adding Biotin-dUTP or FITC-dUTP (Fermentas) in a molar ratio of 1:8 to dTTP in the enzymatic synthesis of poly(dA-dT) as described23. Anthrax lethal toxin was from List Biologicals (5 µg·ml−1 PA and 5 µg·ml−1 LF). Anti-human AIM2 antibody (3B10) was from R. Johnstone (The Peter MacCallum Cancer Institute, Victoria, Australia).
Mice
NLRP3 −/− and Pycard −/− mice were as previously described24. Both strains, as well as NLRP6−/− and NLRP12−/− mice were from Millennium Pharmaceuticals (Cambridge, MA) and Caspase-1-deficient mice were from R. Flavell (Yale University, New Haven, CT). C57BL/6 mice, 129/Sv mice, C57/Bl6 × 129 F1 mice, IL-1R-deficient (Il1r1 −/−) were from Jackson Laboratories (Bar Harbor, ME). All mouse strains were bred and maintained under specific pathogen-free conditions in the animal facilities at the University of Massachusetts Medical School.
Sequence Analysis
Pyrin domain containing sequences were retrieved from UniProt after identifying them in SMART. A multiple sequence alignment of selected pyrin domains was generated using MUSCLE27. Secondary structure elements from the POP1 crystal structure (PDB code: 2HM2) were mapped to the multiple sequence alignment in PFAAT28. Prediction of nuclear targeting sequences and subcellular localization was done using PSORTII (http://psort.ims.u-tokyo.ac.jp/).
Cell culture and stimulation
Bone marrow derived macrophages were generated and cultured in DMEM medium as described26. THP1 cells and macrophage cell lines were cultured as previously described. One day prior to stimulation, THP1 cells were differentiated using 0.5 µM PMA7. ATP (5 mM) was added one hour prior to harvesting supernatants. Poly(dA-dT) DNA and all other DNAs were transfected using Lipofectamine 2000 at a concentration of 1 µg/ml according to the manufacturer’s instructions. Expression plasmids were transfected into 293T cells using GeneJuice (Novagen). Vaccinia virus (WR strain) was used for infection at an MOI of 5 if not indicated otherwise.
ELISA
Cell culture supernatants were assayed for IL-1β by ELISA (BD Biosciences, Franklin Lakes, NJ). To measure intracellular IL-1β, cells were washed and subjected to three freeze thaw cycles in assay diluent.
Confocal microscopy
Confocal microscopy was performed on a Leica SP2 AOBS confocal laser scanning microscope. Separation of CFP and YFP was performed using sequential scanning and 405 and 514 nm excitation.
Flow cytometry Fluorescence Resonance Energy Transfer (FRET)
Three fluorescent intensities were measured: I1 – direct excitation and emission of donor, I2 – indirect excitation and direct emission of acceptor and I3 – direct excitation and emission of acceptor. After full correction for spectral bleed-through and cross excitation, FRET efficiency was calculated on a cell-by-cell basis25 and then FRET efficiency histograms plotted with GraphPad Prism 5.01 (GraphPad Software).
Immunoblot analysis
Immunoblotting was conducted as previously described7 using anti murine caspase-1 p10 (sc-514, Santa Cruz Biotechnology), anti FLAG (M2, Sigma), anti HA (Roche Applied Science), anti CFP (Santa Cruz Biotechnology), anti ASC (Alexis, AL177), anti IFI16 (sc-8023, Santa Cruz Biotechnology) or anti AIM2 (3B10 mouse IgG129).
Co-immunoprecipitation assays
293T cells (24 wells) were transfected with 1550 ng of the PYD-CFP expression plasmids and 50 ng of ASC-HA. 24h later, cell lysates were cleared by centrifugation (20.000 g, 30 min) and subsequently incubated with anti-HA agarose beads for 2h at 4°C. Differentiated THP1 cells were primed with Sendai virus (300 HAU/ml) overnight and subsequently lysed in high salt lysis buffer (250 mM NaCl, 10 mM Tris HCl pH 7.4, 1% CHAPS, protease inhibitor cocktail). Lysates were cleared by centrifugation, salt concentration adjusted to 125 mM NaCl and ASC immunopreciptated using polyclonal rabbit anti-ASC. After 6 washes in both cases beads were boiled with Laemmli buffer for immunoblot analysis.
Quantitative real-time PCR
Quantitative RT-PCR analysis was performed as described26. Primer sequences for murine HPRT1, IFNβ, AIM2 and IL-6 are available upon request.
shRNA mediated silencing
The lentiviral shRNA expression plasmids were from OpenBiosystems (Huntsville, AL). The shRNAs targeting AIM2 are TRCN0000096104 (#1), TRCN0000096105 (#2), TRCN0000096106 (#3). The control shRNA is directed against murine IFIH1 (TRCN0000103648) and was confirmed not to have any impact on NLRP3 or AIM2 expression. The production of viral particles and transduction of target cells was conducted as described on http://www.broad.mit.edu/genome_bio/trc/publicProtocols.html.
AlphaScreen Assay
The AlphaScreen was set up as an association assay. Proteins were transiently expressed in 293T cells and purified using FLAG beads (Sigma) binding to the C-terminal FLAG/HIS tag. The protein of interest was incubated at a concentration of 100 nM with biotinylated ligand at the indicated concentration in PBS, 0.1% BSA, and 0.01% Tween 20 for 60 min. Subsequently, Nickel chelate acceptor beads (binding to the HIS tag) and streptavidin-coated donor beads were added. After 30 min incubation at 25°C in the dark, samples were read in proxiplates with the Envision HT microplate reader (all Perkin Elmer). Data were analyzed by GraphPad Prism.
Reporter assays
All reporter gene assays were conducted as described26.
siRNA transfection
Differentiated THP1 cells were plated at 2.5 × 10E4 cells/ in 96-well plates and transfected with siRNA targeting human AIM2 (sense strand: 5’-CCCGAAGATCAACACGCTTCA-3’), human ASC (sense strand: 5’-CGGGAAGGTCCTGACGGATGA-3’) or human TLR8 (5’-GGGAGUUACUGCUUGAAG A-3’) using 275 ng siRNA and 0.75 µl Lipofectamine 2000. 48h after transfection cells were stimulated as indicated.
Cell viability assay
To quantify cell viability, macrophages (1 × 10E5 cells/ in 96-well plates) were treated as described. After 24h, cells were washed with PBS and incubated in PBS with 5 µM calcein AM (Invitrogen) for 30 min at 37°C. The number of viable cells was assessed by counting fluorescent cells in two independent visual fields (20x) using ImageJ or by determining the overall fluorescence intensity using an Envision HT microplate reader.
Supplementary Material
1
Acknowledgements
The authors would like to thank Anna Cerny for animal husbandry and genotyping and R. Johnstone (The Peter MacCallum Cancer Institute, Victoria, Australia) for the anti-AIM2 antibody. V.H. is supported by a fellowship from the DFG (Ho2783/2–1), E.L. and K.A.F. are supported by grants from the NIH (AI-065483 (to E.L.) and AI-067497 (to K.A.F.).
Footnotes
Author Information: Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
References
- 1.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442(7098):39. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 2.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7(1):40. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 3.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24(1):93. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 5.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451(7179):725. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 6.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452(7183):103. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 7.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9(8):847. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Experimental cell research. 2005;308(1):1. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 9.Ishii, Ken J, Akira, Shizuo Innate immune recognition of, and regulation by, DNA. Trends in Immunology. 2006;27(11):525. doi: 10.1016/j.it.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Manji GA, et al. PYPAF1, a PYRIN-containing Apaf1-like protein that assembles with ASC and regulates activation of NF-kappa B. The Journal of biological chemistry. 2002;277(13):11570. doi: 10.1074/jbc.M112208200. [DOI] [PubMed] [Google Scholar]
- 11.Grenier JM, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett. 2002;530((1–3)):73. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, et al. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. The Journal of biological chemistry. 2002;277(33):29874. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- 13.Finn R, Griffiths-Jones S, Bateman A. Identifying protein domains with the Pfam database. Chapter 2. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis … [et al. 2003 doi: 10.1002/0471250953.bi0205s01. Unit 2 5. [DOI] [PubMed] [Google Scholar]
- 14.Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochemical and biophysical research communications. 2005;327(3):679. doi: 10.1016/j.bbrc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- 15.Landolfo S, Gariglio M, Gribaudo G, Lembo D. The Ifi 200 genes: an emerging family of IFN-inducible genes. Biochimie. 1998;80((8–9)):721. doi: 10.1016/s0300-9084(99)80025-x. [DOI] [PubMed] [Google Scholar]
- 16.Ding Y, et al. Antitumor activity of IFIX, a novel interferon-inducible HIN-200 gene, in breast cancer. Oncogene. 2004;23(26):4556. doi: 10.1038/sj.onc.1207592. [DOI] [PubMed] [Google Scholar]
- 17.Trapani JA, et al. A novel gene constitutively expressed in human lymphoid cells is inducible with interferon-gamma in myeloid cells. Immunogenetics. 1992;36(6):369. doi: 10.1007/BF00218044. [DOI] [PubMed] [Google Scholar]
- 18.Burrus GR, Briggs JA, Briggs RC. Characterization of the human myeloid cell nuclear differentiation antigen: relationship to interferon-inducible proteins. Journal of cellular biochemistry. 1992;48(2):190. doi: 10.1002/jcb.240480210. [DOI] [PubMed] [Google Scholar]
- 19.DeYoung KL, et al. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15(4):453. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 20.Yu JW, et al. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell death and differentiation. 2006;13(2):236. doi: 10.1038/sj.cdd.4401734. [DOI] [PubMed] [Google Scholar]
- 21.Henry T, et al. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. The Journal of experimental medicine. 2007;204(5):987. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun KH, Yu CL, Tang SJ, Sun GH. Monoclonal anti-double-stranded DNA autoantibody stimulates the expression and release of IL-1beta, IL-6, IL-8, IL-10 and TNF-alpha from normal human mononuclear cells involving in the lupus pathogenesis. Immunology. 2000;99(3):352. doi: 10.1046/j.1365-2567.2000.00970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schachman HK, et al. Enzymatic synthesis of deoxyribonucleic acid. VII. Synthesis of a polymer of deoxyadenylate and deoxythymidylate. The Journal of biological chemistry. 1960;235:3242. [PubMed] [Google Scholar]
- 24.Kanneganti TD, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. The Journal of biological chemistry. 2006;281(48):36560. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 25.Szentesi G, et al. Computer program for determining fluorescence resonance energy transfer efficiency from flow cytometric data on a cell-by-cell basis. Computer methods and programs in biomedicine. 2004;75(3):201. doi: 10.1016/j.cmpb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Severa M, Coccia EM, Fitzgerald KA. Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. The Journal of biological chemistry. 2006;281(36):26188. doi: 10.1074/jbc.M604516200. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32(5):1792. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caffrey DR, et al. PFAAT version 2.0: a tool for editing, annotating, and analyzing multiple sequence alignments. BMC bioinformatics. 2007;8:381. doi: 10.1186/1471-2105-8-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cresswell KS, et al. Biochemical and growth regulatory activities of the HIN-200 family member and putative tumor suppressor protein, AIM2. Biochemical and biophysical research communications. 2005;326(2):417. doi: 10.1016/j.bbrc.2004.11.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1