CD2 distinguishes two subsets of human plasmacytoid dendritic cells with distinct phenotype and functions (original) (raw)
. Author manuscript; available in PMC: 2010 Jun 1.
Published in final edited form as: J Immunol. 2009 Jun 1;182(11):6815–6823. doi: 10.4049/jimmunol.0802008
SUMMARY
Plasmacytoid dendritic cells (pDCs) are key regulators of anti-viral immunity. They rapidly secrete IFN-α and cross-present viral antigens thereby launching adaptive immunity. Here we show that activated human pDCs inhibit replication of cancer cells, and kill them in a contact dependent fashion. Expression of CD2 distinguishes two pDC subsets with distinct phenotype and function. Both subsets secrete IFN-α and express Granzyme B and TRAIL. CD2high pDCs uniquely express lysozyme and can be found in tonsils and in tumors. Both subsets launch recall T cell response. However, CD2high pDCs secrete higher levels of IL12 p40, express higher levels of co-stimulatory molecule CD80 and are more efficient in triggering proliferation of naïve allogeneic T cells. Thus, human blood pDCs are composed of subsets with specific phenotype and functions.
Keywords: dendritic cells, T cells, cancer, viruses
INTRODUCTION
Dendritic cells (DCs) represent a complex system of cells with different anatomical localization, distinct subsets, and distinct functions(1, 2). Two main DC pathways bearing common and unique functions have been recognized, i.e. myeloid DCs and plasmacytoid DCs(2, 3). The plasmacytoid DCs (pDCs) have been described independently in the late 70's as plasmacytoid monocytes and plasmacytoid T cells. In the 90's pDCs have been identified as DCs (4, 5) and as the natural IFN producing cells (NIPCs) (6-8). Indeed, upon viral encounter pDCs rapidly secrete considerable amounts of type I Interferons(6, 7) as well as numerous chemokines (9). pDCs can be activated by i) viruses (6, 7); ii) IL-3 and CD40 ligand (IL-3/CD40-L) (5, 10), possibly originating from mast cell triggered for example by parasites; and iii) by bacterial components in the form of CpG DNA (11, 12). Once activated, pDCs induce the expansion and differentiation of antigen-specific memory B and T lymphocytes, yielding plasma cells (13), and cytolytic T cells (CTLs) (14, 15). There is evidence that plasmacytoid DCs are involved in tolerance induction (16-18). pDCs stimulated via TLR or CD40 induce IL-10-secreting regulatory CD4+ T cells owing to the expression of ICOS ligand (19) as well as suppressor CD8+ T cells (20). Finally, pDCs activated via IL3/CD40L can also drive type 2 polarization of CD4+ T cells (10).
pDCs play a key role in disease pathogenesis. Their alteration appears central to immune dysfunction underlying systemic lupus erythematosus (SLE), a prototype autoimmune disease characterized by a break of tolerance to self antigens (21-23). pDC accumulate in skin lesions of SLE (24) where they become activated and secrete type I interferon thereby contributing to tissue damage. Early studies demonstrated that immune complexes are potent stimuli for IFN-α secretion by pDCs in an Fc receptor (CD32)-dependent manner (25, 26). Indeed, chromatin and/or ribonucleoprotein-containing immune complexes can be internalized by pDCs via FcγRIIa, reach the endosomal compartment and activate IFN-α secretion through TLR9 and/or 7 dependent pathways (27, 28). Sera from SLE patients can also induce IFN-α secretion in a TLR7/8-dependent manner (29). The aberrantly produced IFNs are major effectors in the pathogenesis of autoimmunity, mainly by inducing an unabated maturation of peripheral mDCs (22) that can stimulate autoreactive T cells. DNA-containing immune complexes can also activate pDCs to produce IFN-α via high mobility group box 1 protein (HMGB1) activation of TLR9 through a mechanism involving the immunoglobulin superfamily member RAGE, receptor for HMGB1 (30). Similar observations were made in psoriasis (31). There, antimicrobial peptide LL37 (also known as CAMP) has been shown as the key factor that mediates pDC activation (32). LL37 converts inert self-DNA into a potent trigger of interferon production by binding the DNA to form aggregated and condensed structures that are delivered to and retained within early endocytic compartments in pDCs to trigger TLR9 (32). Accumulation of pDCs has also been found in cancer, most particularly in breast cancer, where it has been suggested to represent an independent prognostic factor associated with poor outcomes as measured by overall survival and time to disease progression (33). Furthermore, in vivo signaling via TLR ligands such as CpG (TLR9) (34) or imiquimod (TLR7) (35) leads to tumor regression both in mice and humans possibly via mobilization of pDC. These observations raise a question as to the role of pDCs in tumor microenvironment. The analysis of pDC transcripts revealed several unusual features including the presence of granzyme B (36), an effector molecule of CTLs and NK cells. We show here that pDCs display cytotoxic properties and are composed of subsets with distinct phenotype and functions.
Materials and Methods
Isolation of dendritic cells
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats by Ficoll centrifugation. Monocytes, T cells, B cells and red blood cells were depleted using respective monoclonal antibodies coupled to microbeads (CD3, CD14, CD19, and glycophorin A; Miltenyi Biotec, Auburn, CA). Enriched PBMCs were labeled with CD11c-APC, HLA-DR-QR, CD123-PE and a cocktail of FITC or PE-coupled mAbs against lineage markers: CD3, CD14, CD16, CD19, CD20, CD56 (BD, Franklin Lakes, NJ). In some experiment, cells were also stained additionally with CD2-PE-Cy7 (39C1.5; Beckman Coulter, Fullerton, CA). Alternatively, pDCs were sorted using lineage cocktail, HLA-DR, BDCA-2 (AC144, Miltenyi Biotec) and CD2 (S5.2, BD). Cells were sorted using either FACSVantage or FACSAria (BD). NK cells were sorted from non-depleted PBMCs using CD16-FITC and CD56-PE (BD).
Cell culture
Cultures were established in endotoxin-free medium consisting of RPMI 1640 supplemented with 10% (v/v) heat-inactivated fetal calf serum (FCS), 10mM HEPES, 2mM L-glutamine, penicillin and streptomycin. Sorted total pDCs, mDCs and NK cells were seeded at 5×105 cells/ml in medium containing 20 ng/ml IL-3 (R&D Systems, Minneapolis, MN) and 200 ng/ml CD40L (R&D), 6 μg/ml CpG (InvivoGen, San Diego, CA) or 1 hemagglutination unit/culture well (corresponding to approximately 104 viral particles) Influenza A/PR8/34 virus (Charles River Laboratories, Wilmington, MA). Sorted CD2high and CD2low pDCs were seeded at 1.25×105 cells/ml unless otherwise indicated. After 18-24 hr culture, cells and supernatants were harvested for further analysis.
ELISA
Secretion of Granzyme B was determined using either 1) a sandwich ELISA kit (Research Diagnostic Inc., Flanders NJ) following the manufacturer's protocol; or 2) Luminex assay. Secretion of TRAIL was determined using ELISA kit (Research Diagnostic Inc.).
Cytokine Multiplex Analysis
Cell culture supernatants from activated pDCs were diluted 1:2.5 with culture medium and analyzed using the Beadlyte cytokine assay kit (Millipore, Billerica, MA) as per manufacturer's protocol. Assay plates were run on the Bio-Plex Luminex 100 XYP instrument (Bio-Rad, Hercules, CA) for analysis. Cytokine concentrations were calculated using Bio-Plex Manager 3.0 software with a 5 parameter curve fitting algorithm applied for standard curve calculations.
Microarrays
Total RNA was extracted from sorted pDC subsets of two different normal donors. After two rounds of linear amplification, cRNA was hybridized on Affymetrix HG-U133A GeneChip® arrays. Specific hybridization intensity, probe set signal values and present/absent calls were computed using the GeneChip Operating Software v1.0.0.046. Normalization of signal values per chip was achieved using the GeneChip Operating Software v1.0.0.046 global method of scaling to the target intensity value of 500 per GeneChip®. Gene expression was analyzed with GeneSpring 7.1 software. Transcripts were ranked based on fold change and absolute difference in expression between the two study conditions. Each probe set was previously normalized with a per chip normalization to the 50th percentile, and a per gene normalization to the median of each gene. (This per chip normalization was applied to these data.)
(GEO accession number GSE15215 (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE15215)).
51Cr release assay
Harvested total pDCs, mDCs, NK cells and CD2high, CD2low pDCs were used as effectors. Effectors were co-cultured with 51Cr-labeled K562 at 100:1 E:T ratio or 30:1 E:T ratio. After 18 hr culture, supernatants were analyzed. Percentage of specific lysis was calculated as (cpmexperiment − cpmspontaneous release)/(cpmmaximum release − cpmspontaneous release).
Confocal Imaging
5×105 cells were washed twice with ice cold PBS + 1% BSA, fixed for 20 min in 4% PFA and permeabilized with PBS / 0.05% saponin / 1%BSA. Endogenous biotin was blocked using endogenous biotin blocking reagent (Invitrogen, Carlsban, CA). Cells were then stained with FITC conjugated anti-lysozyme (Research Diagnostic Inc.) and either biotin conjugated anti-BDCA2 (Miltenyi Biotec) or anti-Granzyme B (BD) followed by treatment with either Alexa 568 labeled streptavidin or goat-anti-mouse IgG2a respectively (Invitrogen). Washed cells were then mounted on to slides in VECTASHIELD (Vector Labs, Burlingame, CA) containing TOPRO-2 (Invitrogen). Cells were then imaged using a Leica TCS-NT/SP1 Confocal Microscope equipped with 3 lasers: Ar, Kr, and HeNe, capable of 3 fluorescence channels and transmitted light detection. Images were acquired on a Leica DMIRBE microscope with the 63x Plan APO objective, using Leica TCS v 2.1 software.
Live Cell Microscopy
Activated CD2high or CD2low pDCs were mixed on ice with K562 cells in RPMI medium with 10% human AB serum at a ratio of 2:1 in flat bottom 48-well plates and allowed to settle on ice for 10 min. Plates were then transferred to a 37°C, 5% CO2 live cell imaging chamber. Cells were imaged either in bright field or 488nm florescence every 10 seconds for 8 hr using an Olympus IX-70 microscope at 10x magnification (Olympus 10x PlanFl NA 0.30). Time course images were assembled into stacks using MetaMorph v.6.2r4 (Universal Imaging). For presentation purposes, Quicktime (Apple Computer) movies were generated from assembled stacks. For the purpose of size compression, every tenth frame was inserted into the movie and displayed at a frame rate of 1 frame every 1/10 second, for a total acceleration of ∼1600x. The percentage of pDCs interacting with K562 cells at any given time was determined through an analysis of time course image stacks. Total cells in the frame at a given time point as well as cells interacting with K562 cells were quantitated the Manual Object Count Tool in the MetaMorph software package. Frames before and after any given time point were used as a reference to identify cells as they entered clusters. Percent interaction is expressed as the ratio of bond pDCs over the total number of pDCs in the frame for that time point. To monitor tumor cell killing, K562 cells were labeled with calcein-AM (Invitrogen), washed and cocultured with CD2high pDCs. Killing was assessed by loss of calcein-AM florescence over time by live cell florescence microscopy.
Electron Microscopy
Cells were fixed in 2.5% glutaraldehyde in 0.1M Na cacodylate pH 7.4 for 1 hr at room temperature, washed in cacodylate buffer and postfixed in 1% osmium tetroxide in the same buffer, for 1 hr at RT. Cells were stained “en bloc” in 2% uranyl acetate in 50 mM Na maleate pH 5.2 for 1 hr at RT, then dehydrated in ethanol and embedded in EMbed 812 epoxy resin (all EM reagents from Electron Microscopy Sciences, Fort Washington, PA). Ultrathin sections were cut on a Reichert Ultracut E microtome and collected on formvar- and carbon-coated nickel grids. Sections were stained with 1% lead citrate followed by 2% uranyl acetate, and examined in a Tecnai 12 Biotwin electron microscope (FEI Company). Digital images were recorded using a Morada CCD camera (Olympus Soft Imaging Solutions).
Thymidine Incorporation Assay
Harvested total pDCs and mDCs were used as effectors. Effectors were co-cultured with K562 cells for 18 hr at a ratio of 100:1 E:T ratio. After 18 hr, cell were pulsed with 1 μCi/well of 3H-thymidine for 3 days.. After 3 days, co-cultures were harvested for the incoporporation of 3H-thymidine with liquid scintillation counter (PerkinElmer, Waltham, MA).
Statistical analysis
Non-parametric Mann-Whitney test and paired Wilcoxon test was used as indicated.
Results
Activated blood pDCs display cytotoxic properties
pDCs and mDCs were sorted from peripheral blood mononuclear cells (PBMCs) as LINnegHLA-DR+CD11c−CD123+ cells (pDCs) or LINnegHLA-DR+CD11c+CD123− cells (mDCs) as described earlier (9). pDCs were analyzed either fresh or after overnight activation with three well characterized activators: IL3/CD40 ligand, influenza virus (PR8, H1N1) or CpG A. Freshly isolated pDCs, but not mDCs, show transcription of Granzyme B consistent with earlier studies (36) at levels similar to those observed in cultured antigen-specific CD8+ T cells (not shown). However, while CD8+ T cells show transcription of at least four genes coding different types of Granzyme (37), pDCs express uniquely Granzyme B (not shown). Resting pDCs express the Granzyme B protein (data not shown) and its expression and secretion to culture supernatant is considerably increased after activation (not shown).
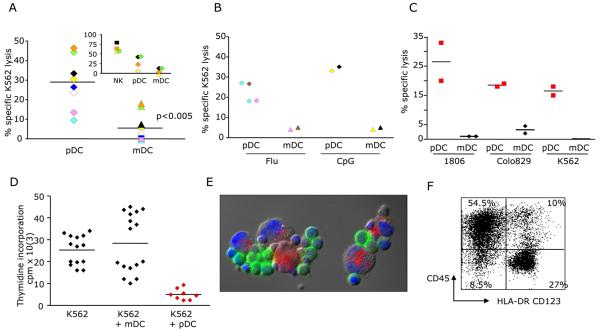
Freshly isolated blood pDCs do not kill efficiently chronic myeloid leukemia derived K562 cells (not shown). However, pDCs activated with IL3/CD40 ligand kill K562 cells as measured by the release of 51Cr (mean ± SEM = 29 ± 5% specific lysis, range 10-46%, n=8; Fig. 1a). Specific lysis in co-cultures of K562 cells with pDCs was significantly higher than that observed with mDCs (mean ± SEM = 7 ± 2.4% specific lysis, range 0-17%; p<0.005; Fig. 1a). The killing of K562 cells by pDCs required high effector: target ratio (E:T ratio=100:1) and long effector: target interaction (18hrs). pDCs also demonstrated cytotoxic function against cancer cells when activated with Influenza virus (mean ± SEM = 22.5 ± 2.5% specific lysis, n=4) or CpG (Fig. 1b). CpG activated pDCs were capable of killing multiple tumor derived cell lines including K562 cells, breast cancer cells (1806 cells) and melanoma cells (Colo829 cells) (Fig. 1b and c). Furthermore, tumor cells co-cultured with activated pDCs showed a significant (p<0.0001) decrease in thymidine incorporation (mean ± SEM = 25.4 ×103 ± 1.8 × 103 cpm vs 5.1 ×103 ± 0.89 × 103 cpm for K562 cells cultured without or with activated pDCs, respectively; Fig. 1d). These results indicate that activated pDCs can halt the growth of tumor cells in vitro. Although the killing of target cells by pDC as measured in a chromium release assay appears not very efficient (high E:T ratio and long incubation time), pDCs are able to abrogate tumor cell growth almost completely (Fig. 1d). These results suggest that pDC might be important in the control of tumor growth.
Figure 1. pDCs kill malignant cells.
(A-B) Killing of K562 cells analyzed in an 18hrs 51Cr release assay with pDCs activated with IL-3CD&40-L (A), Influenza virus, or CpG (B). Specific lysis (ordinate). Paired Wilcoxon test. Color code indicates experiments with cells from the same donor. Insert plot shows killing by NK cells isolated from the same donors. (C) Killing of breast cancer (1806) and melanoma (Colo8290) cells by CpG activated pDC and mDC; 18hrs 51Cr release assay; specific lysis (ordinate). Duplicate wells from one experiment; representative of two experiments. (D) Inhibition of malignant cell replication on 3-day co-culture. Thymidine incorporation (ordinate). Duplicate (or triplicate) wells from four different experiments. (E) Fluorescence microscopy analysis of conjugates between CD45PE labeled K562 cells (red) and CD123FITC/HLA-DRFITC labeled pDCs (green) (1:2 ratio; 45 min). (F) Flow cytometry analysis of conjugate formation between CD45PE labeled K562 cells (red) and CD123FITC/HLA-DRFITC labeled pDCs (green) (1:2 ratio; 45 min) (upper right quadrant).
CD2 distinguishes two fractions of blood pDCs
The killing of K562 by activated pDCs could be blocked by EGTA indicating Ca++ dependence (not shown). Supernatants from activated pDCs or from pDC/K562 cocultures did not kill K562 cells indicating the cell contact dependence (not shown). Accordingly, live microscopy demonstrated that activated pDCs form tight clusters with K562 within 45min of coculture (Fig. 1e). However, not all pDCs appeared bound with K562 cells suggesting the presence of two subsets.
Accordingly, flow cytometric conjugate formation assay, where CD45PE labeled K562 cells are co-incubated with CD123FITC/HLA-DRFITC labeled activated pDCs, showed that only a fraction of pDCs (27% in the experiment displayed in Fig. 1f) were engaged in conjugates with target cells.
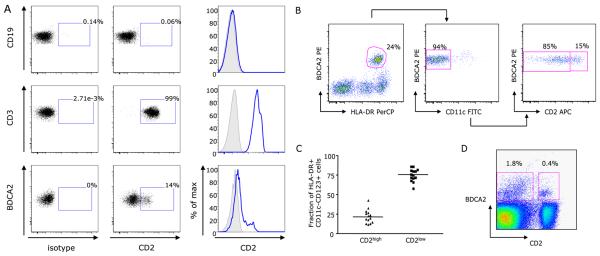
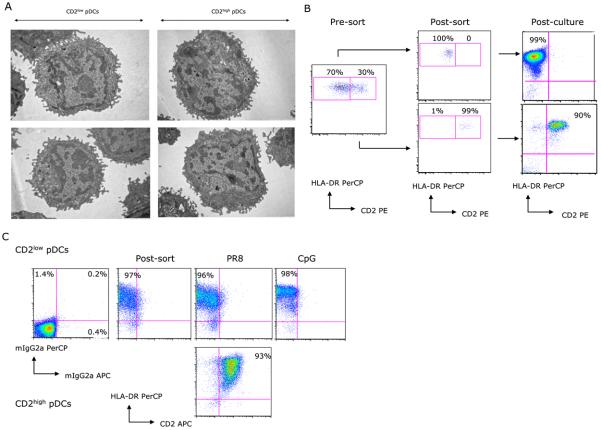
An extensive analysis of T cell and NK cell markers on pDCs defined as lineageneg HLA-DR+ CD11cneg CD123+ (not shown) or as HLA-DR+ CD11cneg BDCA-2+ (Fig. 2a and b) revealed differential expression of CD2 (38-41). In PBMC staining, CD2high pDCs show CD2 staining intensity within the range of that in T cells (Fig. 2a). Analysis of 14 healthy donors showed that 12%-43% of blood pDCs express CD2 at high levels (mean ± SD: 22.5% ± 8.7%) (Fig. 2c). Given that pDCs represent ∼0.25% ± 0.1% of adult peripheral blood mononuclear cells, CD2high pDCs represent about 0.05% of mononuclear cells. CD2high pDCs were also detected in cell suspensions prepared from tonsils as BDCA-2+CD2high double positive cells (mean: 0.77% of cell suspension, range: 0.36% - 1.54%; mean: 22.8% of BDCA-2+ pDCs, range: 16.7% - 32%; Fig. 2d). Giemsa staining of sorted CD2high pDCs revealed morphology characteristic of pDCs (not shown). Similarly, both subsets display pDC ultrastructure in electron microscopy with rich endoplasmic reticulum (Fig. 3a). Furthermore, after sort and overnight culture with IL3/CD40L or with CpG A, CpG B, C or live Influenza virus CD2high pDCs retain the expression of CD2 while CD2low pDC do not acquire expression of CD2 (Fig. 3b) suggesting that CD2 on pDC is not a mere marker of cell activation.
Figure 2. CD2 expression discriminates two subsets of pDCs.
(A) PBMCs staining shows the lack of CD2 expression by blood CD19+ B cells (upper plots), expected peak of expression on CD3+ T cells (middle plots) and a subset of BDCA2+ pDC that show CD2 staining within the range of staining on T cells (lower plots). Representative of the analysis of PBMCs from three donors. (B) pDC subsets distinguished by CD2 expression can also be defined among HLA-DR+BDCA2+ (left panel) CD11cneg (middle panel) cells in pre-enriched PBMCs. (C) Frequency of two subsets within the total pDCs population. (D) BDCA-2+CD2+ pDCs in single cell suspension prepared from tonsil; representative of three independent experiments.
Figure 3. CD2high pDCs and CD2low pDCs show similar ultrastructure.
(A) CD2low (left panels) and CD2high (right panels) pDCs were fixed and processed for transmission electron microscopy. Note the abundance of endoplasmic reticulum in both subsets of cells (arrows). N: nucleus. The bar: 2 micrometers. (B) CD2 staining in flow cytometry on pDC subsets pre-sort, after sort and after 18hrs activation with CpG. (C) Sorted CD2low (upper panels) and CD2high (lower panels) were cultured overnight at 30,000 cells /well with either live PR8 virus 1HA unit/well or CpG 6 μg/ml. Harvested cells were analyzed by flow cytometry for their surface expression of CD2. Upper panels: CD2low pDCs isotype control, CD2 staining after sort, CD2 staining after PR8 exposure and CD2 staining after CpG exposure. Lower panels: CD2high pDCs after PR8 exposure. Representative experiment of four performed testing different conditions of pDC activation.
Thus, pDC can be separated according to CD2 expression, and CD2high and CD2low pDCs display comparable morphology and ultrastructure.
CD2high pDCs express lysozyme
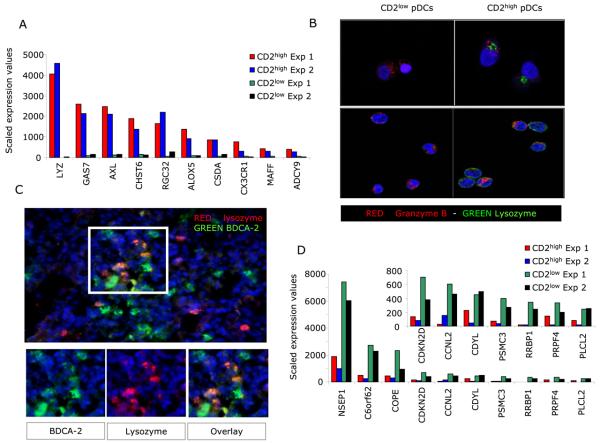
A microarray analysis of sorted CD2high and CD2low subsets of pDCs revealed that, the most significantly over-expressed transcript in CD2high pDCs was lysozyme (139 fold over-expression). This is unexpected as lysozyme is known to be a product of myeloid cells such as neutrophils, monocytes as well as of epithelial cells, and play a role in the breakdown of bacterial cell walls (42) (Fig. 4a). Accordingly, CD2high pDCs exclusively expressed lysozyme in their intracellular compartments (Fig. 4b), thus confirming the existence of two distinct pDC subsets. However, adding lysozyme to cocultures of CD2low pDCs and K562 cells was not sufficient to reproduce K562 cell killing (data not shown). Interestingly, cells co-staining with anti-BDCA-2 and anti-lysozyme antibodies could be observed in biopsies of melanoma tumors from patients (Fig. 4c). Transcription of several other molecules could distinguish the two subsets (Fig. 4a and d). Beside lysozyme, the top five transcripts over-expressed in CD2high pDCs included Gas7, a member of growth arrest-specific genes expressed in neurons (43) and possibly involved in dendrite formation (44); AXL, a member of Tyro 3 family of receptor tyrosine kinases with anti-inflammatory activities (45) that is involved in type I IFN mediated suppression of TNF production (46); CHST6, a gene encoding an enzyme carbohydrate sulphotransferase (47); and RGC32, a member of Response Genes to Complement (48), whose expression was found upregulated in blood cells of patients with Hyper IgE syndrome (49) (Fig. 4a). The transcription pattern of CD2low pDCs was strikingly different and the most over-expressed transcript NSEP1 encodes Y box binding protein 1 (YB-1) (50), a transcription factor involved in the regulation of cell cycle (50) as well as in biological effects of IFN-γ (51, 52). Interestingly, YB-1 protein appears to interact with different viruses including HIV (53, 54). Other molecules implicated in cell cycle regulation were also transcribed by CD2low pDCs including CDKN2D, an inhibitor of cyclin dependent kinases (55) and CCNL2, cyclin L2 (56). Finally, the top transcripts also encompassed molecules relevant for endoplasmic reticulum such as COPE (57 ) (Fig. 4d) and RRBP1 ribosome binding protein (58) (not shown).
Figure 4. CD2high pDCs and CD2low pDCs show distinct transcriptional profiles.
(A) Top ten genes overexpressed in CD2high pDCs as compared to CD2low pDCs (determined by fold change). Microarray scaled expression data; two experiments with pDC subsets sorted from two different donors. (B) Sorted pDCs subsets are stained with FITC conjugated anti-Lysozyme mAb and either biotin conjugated anti-BDCA2 or anti-Granzyme-B mAb followed by treatment with either Alexa 568 labeled streptavidin or goat-anti-mouse IgG2a, respectively. Images are acquired on a Leica DMIRBE microscope with the 63x Plan APO objective, using Leica TCS v 2.1 software. (C) staining of melanoma tumor. (D) Top ten genes overexpressed in CD2low pDCs as compared to CD2high pDCs (determined by fold change). Microarray scaled expression data; two experiments with pDC subsets sorted from two different donors. Insert plot shows genes with expression values <1000.
Thus, pDC subsets distinguished by CD2 display distinct and unique transcription and protein expression profiles.
Activated CD2high pDCs form tight clusters with other cells
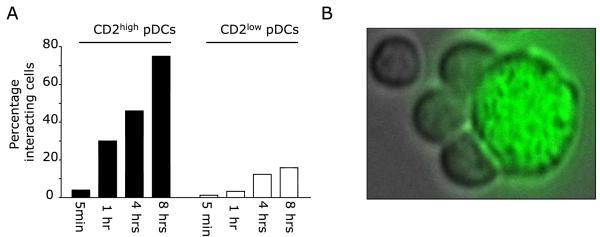
Both pDC subsets responded to activation via CD40 with increased transcription and expression of Granzyme B and TRAIL (not shown). Cell tracking in live microscopy indicated that, after 1hr, approximately 30% of CD2high pDCs, but <10% of CD2low pDCs, were capable of interacting with K562 cells (Fig. 5a, movie Fig. S1). After 8hrs of observation, up to 80% of CD2high pDCs were interacting with K562 cells when <20% of CD2low pDCs interacted with K562 (Fig. 5a). The interactions between CD2high pDCs and K562 cells involved several pDCs bound to a K562 target cell (Fig. 5b), and lead in many instances to K562 cell death (movie Fig. S2). The formation of very tight clusters with other cells might permit pDCs to “screen” their surface and may serve as means to eliminate cells and/or capture antigens for example through nibbling (59) as it has been described for myeloid DCs. Thus, CD2high pDCs could represent a link between the innate and the adaptive immunity in the context of cancer.
Figure 5. CD2high pDCs strongly bind target cells.
(A) Activated CD2high or CD2low pDCs were mixed with K562 cells at 2:1 ratio and monitored in 37°C 5% CO2 live cell imaging chamber. The percentage of pDCs interacting with K562 cells was determined through an analysis of time course image stacks. Percent interaction is expressed as the ratio of bound pDCs over the total number of pDCs in the frame for that time point. (B) K562 cell (green) surrounded by tightly bound CD2+ pDCs.
Activated CD2high pDCs induce proliferation of naïve allogeneic T cells
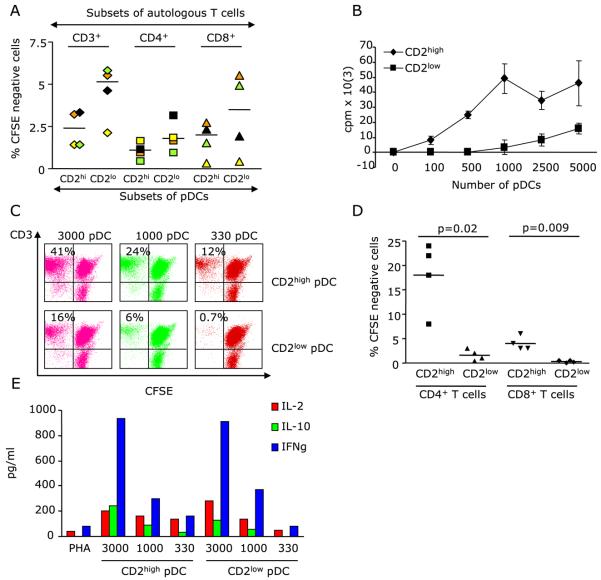
We next analyzed the capacity of CD2high and CD2low pDCs to present antigen to T cells. CD2high and CD2low pDCs were pulsed with influenza virus and cocultured for 6 days with CFSE-labeled autologous CD4+ and CD8+T cells. As shown in Fig. 6a, both subsets were able to trigger proliferation of autologous T cells to influenza virus. To determine their capacity to launch primary immune responses, pDC subsets were activated with CpG and used in standard mixed lymphocyte reaction (MLR). As shown in Fig. 6b, CD2high pDCs were more efficient than CD2low pDCs in the induction of naive allogeneic T cell proliferation as measured by thymidine incorporation. These data were further confirmed by the analysis of T cell division by CFSE dilution (Fig. 6c). The superiority of CD2high pDCs in stimulating MLR was observed with purified CD4+ T cells (mean ± SEM = 18 ± 3.5% vs 1.6 ± 0.6% for CD2high and CD2low pDCs, respectively; p=0.02; Fig. 6d) and with CD8+T cells (mean ± SEM = 4 ± 0.6% vs 0.3 ± 0.1% for CD2high and CD2low pDCs, respectively; p=0.009; Fig. 6d). However, despite their lower proliferation rate, on a per cell basis, T cells exposed to CD2low pDCs produced equivalent levels of IL-2, IL-10 and IFN-γ as those exposed to CD2high pDCs (Fig. 6e).
Figure 6. CD2 expression discriminates two subsets of pDCs one of which is immunogenic.
(A) Sorted pDC subsets are pulsed with heat-inactivated Influenza virus and used in co-cultures with CFSE-labeled autologous T cells. T cell proliferation (CFSE dilution) is measured at day 6. (B-D) Sorted pDC subsets are activated with CpG and plated with naïve allogeneic T cells. T cell proliferation is measured by thymidine incorporation (B; representative experiment of three performed) or by CFSE dilution (C; representative experiment) and (D; four independent experiments). (E) Supernatants of pDC/T cell cocultures are analyzed at day 5 for cytokine secretion.
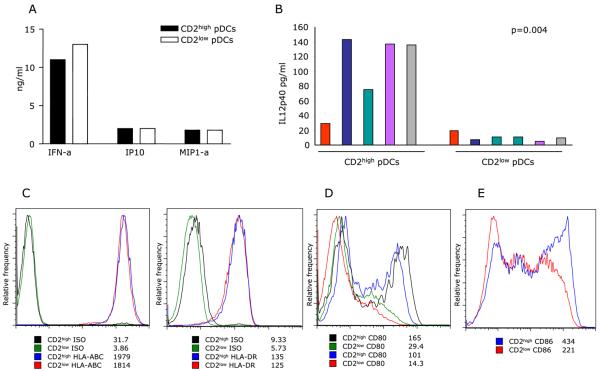
Upon challenge with live Influenza virus or CpG, CD2high pDCs and CD2low pDCs secreted comparable amounts of IFN-α, IP-10 and MIP1-α (Fig. 7a and not shown). Interestingly, CD2high pDCs activated with live Influenza virus secreted higher levels of IL12 p40 than CD2low pDCs isolated from the same donor (Fig. 7b). Similar difference in IL12 p40 secretion could be observed after activation with CpG (data not shown). Flow cytometry analysis demonstrated the same levels of MHC class I or MHC class II molecules expression by the two pDC subsets (Fig. 7c). Both subsets also express CD83 typically found on mature DCs (not shown). Analysis of co-stimulatory molecules expression showed that CD2high pDCs can up-regulate CD80 after activation with CpG while CD2low pDCs do so poorly (Fig. 7d). Expression of CD86 is more variable and both subsets can up-regulate CD86 though CD2high pDCs show higher intensity of staining (Fig. 7e).
Figure 7. Activated CD2high pDCs and CD2low pDCs show unique and distinct features.
(A-B) Cytokine secretion (ng/ml, ordinate) by pDC subsets after activation with Influenza virus (Luminex analysis). (A) Representative of eight experiments; (B) five different donors, color indicates pDC subsets isolated from the same donor; 25,000-30,000 cells per well. (C-E) Flow cytometry analysis of protein expression by activated pDCs subsets: MHC class I and class II molecules (B; two different donors); CD80 (C; two different donors) and CD86 (D; representative of four different donors). Values indicate mean fluorescence intensity.
Thus, upon activation, pDC subsets defined by high expression of CD2 secrete higher levels of IL12 p40, show higher expression of CD80 and a distinct capacity to trigger naïve T cell expansion.
DISCUSSION
The present study demonstrates that human pDCs are composed of two subsets, which can be distinguished by the expression of CD2. These two subsets are found not only in blood but also in tonsils. In addition to their circulation in the blood and secondary lymphoid organs such as tonsils, CD2high pDCs can be detected in some tumor biopsies suggesting their potential involvement in tumor immunosurveillance. Both subsets secrete IFN-α and express the cytotoxic molecules Granzyme B and TRAIL. The CD2high pDCs are potent in initiating T cell immune responses. In contrast, the CD2low pDCs appear to display a limited capacity to induce allogeneic T cell proliferation. These different functional properties of CD2high pDCs and CD2low pDCs are associated to distinct transcription profiles, differential secretion of IL12 p40 and with differential expression of co-stimulatory molecule CD80 on activation.
In concordance with other studies, Granzyme B inhibitor does not seem to affect cytotoxic function of pDC. Granzyme B-mediated killing of target cells seems to be completely dependent on the presence of perforin (60-62), though alternative mechanism has been suggested (63). As pDCs do not express perforin, they must either use another protein that can substitute or they use Granzyme for biological function other than killing. Certain acute viral (HIV, EBV) infections seem associated with elevated plasma levels of Granzyme A and B (64). While it was considered to be associated with an ongoing CTL activation (64), the source of extracellular Granzyme B in these patients may need to be revisited in the view of our results. Furthermore, elevated Granzyme B plasma levels were also observed in patients with parasite infection with Plasmodium falciparum (65) and in severe bacterial infection (66). These in vivo observations are consistent with high secretion triggered in vitro by pDC activation via CD40 and TLR ligands, respectively. Recent studies demonstrate the involvement of Granzyme B in extracellular matrix remodeling via cleavage of vitronectin, laminin and fibronectin (67). That may lead in vitro to anoikis, cell death due to lack of extracellular contact (67). This, together with their capacity to secrete Granzyme B, could point to a novel role of pDC in inflammation as well as tumor development and progression.
Thus, the killing mechanism remains to be established. Three recent studies in the human and in the mouse suggested a role of TRAIL-mediated cytotoxicity exerted by cells with pDC properties (68-70). Our preliminary results corroborate the potential role of TRAIL (not shown) however the inhibition is not complete suggesting contribution of other mechanisms. The killing is unlikely to be due to contaminating NK cells as it requires high E:T target ratio, is slow (NK cell mediated-lysis usually occurs at 4hrs), we do not detect IFN-γ transcription or secretion as well as we do not do detect perforin (data not shown).
Resting CD2high pDCs highly transcribe genes involved in the immune functions such as lysozyme, AXL or fractalkine receptor. Another feature is the expression of two genes linked to Th2 polarization (ALOX5) and Th2 signature such as IgE (RGC32). Thus, CD2high pDCs might be responsible for the capacity of pDCs to induce type 2 immunity (10). Conversely, CD2low pDCs highly transcribe genes involved in cell cycle regulation and in secretory functions. Non-activated CD2high pDC also show 2-fold higher transcription of CD2 as compared to non-activated CD2low pDC (Supplementary Fig. 3). While upon activation both subsets express MHC class molecules and CD83, CD2high pDCs up-regulate CD80. While both subsets secrete similar levels of cytokines and chemokines such as IL-6, IL-8, TNF alpha, IFN alpha, IP-10, MIP1-α, and MIP1-β on activation, CD2high pDCs secrete IL-12 p40. Preliminary results show 2-fold enhanced transcription of p19 in microarrays of activated CD2high pDCs (not shown). Thus, the identification of common and unique transcripts further supports the notion of two pDC subsets in the humans.
The differential expression of lymphoid-related genes (RAG1 and Ig rearrangement products)(71) or proteins (CD4)(72) and Ly49Q(73) has been used to demonstrate the existence of murine pDC subsets. Similar to our results in the human, murine pDC subsets differ in their capacity to trigger allogeneic T cell proliferation and to secrete IL12p70 and IFN-γ (71).
pDCs have been shown to arise from both myeloid and lymphoid committed hematopoietic progenitor cells (CMP and CLP, respectively) (74, 75). The CD2 subset clearly demonstrates the prevalent expression of myeloid related genes thus suggesting that pDCs originating from CMP might have distinct functions than those originating from CLP. Thus, both in human and mouse pDCs constitute a heterogeneous population of cells which display a remarkable array of immune functions. The existence of two pDC subsets might provide a framework for explanation of a diverse array of immune responses elicited by pDCs ranging from CD8+T cell tolerance/anergy, generation of regulatory T cells, to Th2 polarization.
Supplementary Material
Fig S1a-movie
Fig S1b-movie
Fig S2-movie
Fig S3
Acknowledgments
We thank Elizabeth Kraus and Sebastien Coquery for help in flow cytometry and cell sorting; Windy Allman for help with microarrays; Sophie Paczesny for help with analysis of antigen-specific CD8+T cells; Cindy Samuelsen, Carson Harrod and Nicolas Taquet for administrative and facility support; Dr. Gerard Zurawski for critical reading of the manuscript; Dr. Michael Ramsay for continuous support. JB is the recipient of the Caruth Chair for Transplantation Immunology Research. AKP is the recipient of the Michael A. Ramsay Chair for Cancer Immunology Research.
Footnotes
1
Supported by Baylor Health Care Systems Foundation and the National Institutes of Health (U19 AIO57234 and CA78846 to JB).
References
- 1.Steinman RM. The dendritic cell system and its role in immunogenicity. Annu Rev Immunol. 1991;9:271–296. doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu Y, Pulendran B, Palucka K. Immunobiology of dendritic cells. Ann Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 3.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 4.O'Doherty U, Peng M, Gezelter S, Swiggard WJ, Betjes M, Bhardwaj N, Steinman RM. Human blood contains two subsets of dendritic cells, one immunologically mature and the other immature. Immunology. 1994;82:487–493. [PMC free article] [PubMed] [Google Scholar]
- 5.Grouard G, Rissoan MC, Filgueira L, Durand I, Banchereau J, Liu YJ. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J Exp Med. 1997;185:1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, Antonenko S, Liu YJ. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 7.Cella M, Jarrossay D, Facchetti F, Alebardi O, Nakajima H, Lanzavecchia A, Colonna M. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce high levels of type I IFN. Nature Medicine. 1999:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- 8.Asselin-Paturel C, Trinchieri G. Production of type I interferons: plasmacytoid dendritic cells and beyond. J Exp Med. 2005;202:461–465. doi: 10.1084/jem.20051395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piqueras B, Connolly J, Freitas H, Palucka AK, Banchereau J. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood. 2006;107:2613–2618. doi: 10.1182/blood-2005-07-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rissoan MC, Soumelis V, Kadowaki N, Grouard G, Briere F, de Waal Malefyt R, Liu YJ. Reciprocal control of T helper cell and dendritic cell differentiation. Science. 1999;283:1183–1186. doi: 10.1126/science.283.5405.1183. [DOI] [PubMed] [Google Scholar]
- 11.Bauer M, Redecke V, Ellwart JW, Scherer B, Kremer JP, Wagner H, Lipford GB. Bacterial CpG-DNA triggers activation and maturation of human CD11c−, CD123+ dendritic cells. J Immunol. 2001;166:5000–5007. doi: 10.4049/jimmunol.166.8.5000. [DOI] [PubMed] [Google Scholar]
- 12.Krug A, Rothenfusser S, Hornung V, Jahrsdorfer B, Blackwell S, Ballas ZK, Endres S, Krieg AM, Hartmann G. Identification of CpG oligonucleotide sequences with high induction of IFN-alpha/beta in plasmacytoid dendritic cells. Eur J Immunol. 2001;31:2154–2163. doi: 10.1002/1521-4141(200107)31:7<2154::aid-immu2154>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 13.Jego G, Palucka AK, Blanck JP, Chalouni C, Pascual V, Banchereau J. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity. 2003;19:225–234. doi: 10.1016/s1074-7613(03)00208-5. [DOI] [PubMed] [Google Scholar]
- 14.Fonteneau JF, Gilliet M, Larsson M, Dasilva I, Munz C, Liu YJ, Bhardwaj N. Activation of influenza virus-specific CD4+ and CD8+ T cells: a new role for plasmacytoid dendritic cells in adaptive immunity. Blood. 2003;101:3520–3526. doi: 10.1182/blood-2002-10-3063. [DOI] [PubMed] [Google Scholar]
- 15.Di Pucchio T, Chatterjee B, Smed-Sorensen A, Clayton S, Palazzo A, Montes M, Xue Y, Mellman I, Banchereau J, Connolly JE. Direct proteasome-independent cross-presentation of viral antigen by plasmacytoid dendritic cells on major histocompatibility complex class I. Nat Immunol. 2008;9:551–557. doi: 10.1038/ni.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Heer HJ, Hammad H, Soullie T, Hijdra D, Vos N, Willart MA, Hoogsteden HC, Lambrecht BN. Essential role of lung plasmacytoid dendritic cells in preventing asthmatic reactions to harmless inhaled antigen. J Exp Med. 2004;200:89–98. doi: 10.1084/jem.20040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochando JC, Homma C, Yang Y, Hidalgo A, Garin A, Tacke F, Angeli V, Li Y, Boros P, Ding Y, Jessberger R, Trinchieri G, Lira SA, Randolph GJ, Bromberg JS. Alloantigen-presenting plasmacytoid dendritic cells mediate tolerance to vascularized grafts. Nat. Immunol. 2006;7:652–662. doi: 10.1038/ni1333. [DOI] [PubMed] [Google Scholar]
- 18.Merad M, Collin M, Bromberg J. Dendritic cell homeostasis and trafficking in transplantation. Trends Immunol. 2007 doi: 10.1016/j.it.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Ito T, Yang M, Wang YH, Lande R, Gregorio J, Perng OA, Qin XF, Liu YJ, Gilliet M. Plasmacytoid dendritic cells prime IL-10-producing T regulatory cells by inducible costimulator ligand. J Exp Med. 2007;204:105–115. doi: 10.1084/jem.20061660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilliet M, Liu YJ. Generation of human CD8 T regulatory cells by CD40 ligand-activated plasmacytoid dendritic cells. J Exp Med. 2002;195:695–704. doi: 10.1084/jem.20011603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cederblad B, Blomberg S, Vallin H, Perers A, Alm GV, Ronnblom L. Patients with systemic lupus erythematosus have reduced numbers of circulating natural interferon-alpha- producing cells. J Autoimmun. 1998;11:465–470. doi: 10.1006/jaut.1998.0215. [DOI] [PubMed] [Google Scholar]
- 22.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294:1540–1543. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 23.Palucka AK, Blanck JP, Bennett L, Pascual V, Banchereau J. Cross-regulation of TNF and IFN-{alpha} in autoimmune diseases. Proc Natl Acad Sci U S A. 2005;102:3372–3377. doi: 10.1073/pnas.0408506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farkas L, Beiske K, Lund-Johansen F, Brandtzaeg P, Jahnsen FL. Plasmacytoid Dendritic Cells (Natural Interferon- alpha/beta-Producing Cells) Accumulate in Cutaneous Lupus Erythematosus Lesions. Am J Pathol. 2001;159:237–243. doi: 10.1016/s0002-9440(10)61689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bave U, Alm GV, Ronnblom L. The combination of apoptotic U937 cells and lupus IgG is a potent IFN-alpha inducer [In Process Citation] J Immunol. 2000;165:3519–3526. doi: 10.4049/jimmunol.165.6.3519. [DOI] [PubMed] [Google Scholar]
- 26.Bave U, Magnusson M, Eloranta ML, Perers A, Alm GV, Ronnblom L. Fc gamma RIIa is expressed on natural IFN-alpha-producing cells (plasmacytoid dendritic cells) and is required for the IFN-alpha production induced by apoptotic cells combined with lupus IgG. J Immunol. 2003;171:3296–3302. doi: 10.4049/jimmunol.171.6.3296. [DOI] [PubMed] [Google Scholar]
- 27.Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, Chang B, Duramad O, Coffman RL. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 29.Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Invest. 2005;115:407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, Hua J, An LL, Audoly L, La Rosa G, Bierhaus A, Naworth P, Marshak-Rothstein A, Crow MK, Fitzgerald KA, Latz E, Kiener PA, Coyle AJ. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 31.Nestle FO, Conrad C, Tun-Kyi A, Homey B, Gombert M, Boyman O, Burg G, Liu YJ, Gilliet M. Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J Exp Med. 2005;202:135–143. doi: 10.1084/jem.20050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Su B, Nestle FO, Zal T, Mellman I, Schroder JM, Liu YJ, Gilliet M. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 33.Treilleux I, Blay JY, Bendriss-Vermare N, Ray-Coquard I, Bachelot T, Guastalla JP, Bremond A, Goddard S, Pin JJ, Barthelemy-Dubois C, Lebecque S. Dendritic cell infiltration and prognosis of early stage breast cancer. Clin Cancer Res. 2004;10:7466–7474. doi: 10.1158/1078-0432.CCR-04-0684. [DOI] [PubMed] [Google Scholar]
- 34.Krieg AM. Therapeutic potential of Toll-like receptor 9 activation. Nat Rev Drug Discov. 2006;5:471–484. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 35.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 36.Rissoan MC, Duhen T, Bridon JM, Bendriss-Vermare N, Peronne C, de Saint Vis B, Briere F, Bates EE. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood. 2002;100:3295–3303. doi: 10.1182/blood-2002-02-0638. [DOI] [PubMed] [Google Scholar]
- 37.Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol. 2003;3:361–370. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- 38.Res PC, Couwenberg F, Vyth-Dreese FA, Spits H. Expression of pTalpha mRNA in a committed dendritic cell precursor in the human thymus. Blood. 1999;94:2647–2657. [PubMed] [Google Scholar]
- 39.Pulendran B, Banchereau J, Burkeholder S, Kraus E, Guinet E, Chalouni C, Caron D, Maliszewski C, Davoust J, Fay J, Palucka K. Flt3-ligand and granulocyte colony-stimulating factor mobilize distinct human dendritic cell subsets in vivo. J Immunol. 2000;165:566–572. doi: 10.4049/jimmunol.165.1.566. [DOI] [PubMed] [Google Scholar]
- 40.Comeau MR, Van der Vuurst de Vries AR, Maliszewski CR, Galibert L. CD123bright plasmacytoid predendritic cells: progenitors undergoing cell fate conversion? J Immunol. 2002;169:75–83. doi: 10.4049/jimmunol.169.1.75. [DOI] [PubMed] [Google Scholar]
- 41.Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, Buck DW, Schmitz J. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol. 2000;165:6037–6046. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]
- 42.Fleming A. On a remarkable bacteriolytic element found in tissues and secretions. Proc R Soc Lond B Biol Sci. 1922;93:306–317. [Google Scholar]
- 43.Ju YT, Chang AC, She BR, Tsaur ML, Hwang HM, Chao CC, Cohen SN, Lin-Chao S. gas7: A gene expressed preferentially in growth-arrested fibroblasts and terminally differentiated Purkinje neurons affects neurite formation. Proc Natl Acad Sci U S A. 1998;95:11423–11428. doi: 10.1073/pnas.95.19.11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.She BR, Liou GG, Lin-Chao S. Association of the growth-arrest-specific protein Gas7 with F-actin induces reorganization of microfilaments and promotes membrane outgrowth. Exp Cell Res. 2002;273:34–44. doi: 10.1006/excr.2001.5435. [DOI] [PubMed] [Google Scholar]
- 45.Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB, Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131:1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 46.Sharif MN, Sosic D, Rothlin CV, Kelly E, Lemke G, Olson EN, Ivashkiv LB. Twist mediates suppression of inflammation by type I IFNs and Axl. J Exp Med. 2006;203:1891–1901. doi: 10.1084/jem.20051725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akama TO, Nishida K, Nakayama J, Watanabe H, Ozaki K, Nakamura T, Dota A, Kawasaki S, Inoue Y, Maeda N, Yamamoto S, Fujiwara T, Thonar EJ, Shimomura Y, Kinoshita S, Tanigami A, Fukuda MN. Macular corneal dystrophy type I and type II are caused by distinct mutations in a new sulphotransferase gene. Nat Genet. 2000;26:237–241. doi: 10.1038/79987. [DOI] [PubMed] [Google Scholar]
- 48.Badea TC, Niculescu FI, Soane L, Shin ML, Rus H. Molecular cloning and characterization of RGC-32, a novel gene induced by complement activation in oligodendrocytes. J Biol Chem. 1998;273:26977–26981. doi: 10.1074/jbc.273.41.26977. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka T, Takada H, Nomura A, Ohga S, Shibata R, Hara T. Distinct gene expression patterns of peripheral blood cells in hyper-IgE syndrome. Clin Exp Immunol. 2005;140:524–531. doi: 10.1111/j.1365-2249.2005.02805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shnyreva M, Schullery DS, Suzuki H, Higaki Y, Bomsztyk K. Interaction of two multifunctional proteins. Heterogeneous nuclear ribonucleoprotein K and Y-box-binding protein. J Biol Chem. 2000;275:15498–15503. doi: 10.1074/jbc.275.20.15498. [DOI] [PubMed] [Google Scholar]
- 51.Dooley S, Said HM, Gressner AM, Floege J, En-Nia A, Mertens PR. Y-box protein-1 is the crucial mediator of antifibrotic interferon-gamma effects. J Biol Chem. 2006;281:1784–1795. doi: 10.1074/jbc.M510215200. [DOI] [PubMed] [Google Scholar]
- 52.Higashi K, Inagaki Y, Suzuki N, Mitsui S, Mauviel A, Kaneko H, Nakatsuka I. Y-box-binding protein YB-1 mediates transcriptional repression of human alpha 2(I) collagen gene expression by interferon-gamma. J Biol Chem. 2003;278:5156–5162. doi: 10.1074/jbc.M208724200. [DOI] [PubMed] [Google Scholar]
- 53.Safak M, Gallia GL, Ansari SA, Khalili K. Physical and functional interaction between the Y-box binding protein YB-1 and human polyomavirus JC virus large T antigen. J Virol. 1999;73:10146–10157. doi: 10.1128/jvi.73.12.10146-10157.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ansari SA, Safak M, Gallia GL, Sawaya BE, Amini S, Khalili K. Interaction of YB-1 with human immunodeficiency virus type 1 Tat and TAR RNA modulates viral promoter activity. J Gen Virol. 1999;80(Pt 10):2629–2638. doi: 10.1099/0022-1317-80-10-2629. [DOI] [PubMed] [Google Scholar]
- 55.Okuda T, Hirai H, Valentine VA, Shurtleff SA, Kidd VJ, Lahti JM, Sherr CJ, Downing JR. Molecular cloning, expression pattern, and chromosomal localization of human CDKN2D/INK4d, an inhibitor of cyclin D-dependent kinases. Genomics. 1995;29:623–630. doi: 10.1006/geno.1995.9957. [DOI] [PubMed] [Google Scholar]
- 56.Yang L, Li N, Wang C, Yu Y, Yuan L, Zhang M, Cao X. Cyclin L2, a novel RNA polymerase II-associated cyclin, is involved in pre-mRNA splicing and induces apoptosis of human hepatocellular carcinoma cells. J Biol Chem. 2004;279:11639–11648. doi: 10.1074/jbc.M312895200. [DOI] [PubMed] [Google Scholar]
- 57.Shima DT, Scales SJ, Kreis TE, Pepperkok R. Segregation of COPI-rich and anterograde-cargo-rich domains in endoplasmic-reticulum-to-Golgi transport complexes. Curr Biol. 1999;9:821–824. doi: 10.1016/s0960-9822(99)80365-0. [DOI] [PubMed] [Google Scholar]
- 58.Savitz AJ, Meyer DI. Identification of a ribosome receptor in the rough endoplasmic reticulum. Nature. 1990;346:540–544. doi: 10.1038/346540a0. [DOI] [PubMed] [Google Scholar]
- 59.Harshyne LA, Zimmer MI, Watkins SC, Barratt-Boyes SM. A role for class A scavenger receptor in dendritic cell nibbling from live cells. J Immunol. 2003;170:2302–2309. doi: 10.4049/jimmunol.170.5.2302. [DOI] [PubMed] [Google Scholar]
- 60.Catalfamo M, Henkart PA. Perforin and the granule exocytosis cytotoxicity pathway. Curr Opin Immunol. 2003;15:522–527. doi: 10.1016/s0952-7915(03)00114-6. [DOI] [PubMed] [Google Scholar]
- 61.Shi L, Keefe D, Durand E, Feng H, Zhang D, Lieberman J. Granzyme B binds to target cells mostly by charge and must be added at the same time as perforin to trigger apoptosis. J Immunol. 2005;174:5456–5461. doi: 10.4049/jimmunol.174.9.5456. [DOI] [PubMed] [Google Scholar]
- 62.Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Motyka B, Korbutt G, Pinkoski MJ, Heibein JA, Caputo A, Hobman M, Barry M, Shostak I, Sawchuk T, Holmes CF, Gauldie J, Bleackley RC. Mannose 6-phosphate/insulin-like growth factor II receptor is a death receptor for granzyme B during cytotoxic T cell-induced apoptosis. Cell. 2000;103:491–500. doi: 10.1016/s0092-8674(00)00140-9. [DOI] [PubMed] [Google Scholar]
- 64.Spaeny-Dekking EH, Hanna WL, Wolbink AM, Wever PC, Kummer AJ, Swaak AJ, Middeldorp JM, Huisman HG, Froelich CJ, Hack CE. Extracellular granzymes A and B in humans: detection of native species during CTL responses in vitro and in vivo. J Immunol. 1998;160:3610–3616. [PubMed] [Google Scholar]
- 65.Hermsen CC, Konijnenberg Y, Mulder L, Loe C, van Deuren M, van der Meer JW, van Mierlo GJ, Eling WM, Hack CE, Sauerwein RW. Circulating concentrations of soluble granzyme A and B increase during natural and experimental Plasmodium falciparum infections. Clin Exp Immunol. 2003;132:467–472. doi: 10.1046/j.1365-2249.2003.02160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lauw FN, Simpson AJ, Hack CE, Prins JM, Wolbink AM, van Deventer SJ, Chaowagul W, White NJ, van Der Poll T. Soluble granzymes are released during human endotoxemia and in patients with severe infection due to gram-negative bacteria. J Infect Dis. 2000;182:206–213. doi: 10.1086/315642. [DOI] [PubMed] [Google Scholar]
- 67.Buzza MS, Zamurs L, Sun J, Bird CH, Smith AI, Trapani JA, Froelich CJ, Nice EC, Bird PI. Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin. J Biol Chem. 2005;280:23549–23558. doi: 10.1074/jbc.M412001200. [DOI] [PubMed] [Google Scholar]
- 68.Chaperot L, Blum A, Manches O, Lui G, Angel J, Molens JP, Plumas J. Virus or TLR agonists induce TRAIL-mediated cytotoxic activity of plasmacytoid dendritic cells. J Immunol. 2006;176:248–255. doi: 10.4049/jimmunol.176.1.248. [DOI] [PubMed] [Google Scholar]
- 69.Taieb J, Chaput N, Menard C, Apetoh L, Ullrich E, Bonmort M, Pequignot M, Casares N, Terme M, Flament C, Opolon P, Lecluse Y, Metivier D, Tomasello E, Vivier E, Ghiringhelli F, Martin F, Klatzmann D, Poynard T, Tursz T, Raposo G, Yagita H, Ryffel B, Kroemer G, Zitvogel L. A novel dendritic cell subset involved in tumor immunosurveillance. Nat Med. 2006;12:214–219. doi: 10.1038/nm1356. [DOI] [PubMed] [Google Scholar]
- 70.Stary G, Bangert C, Tauber M, Strohal R, Kopp T, Stingl G. Tumoricidal activity of TLR7/8-activated inflammatory dendritic cells. J Exp Med. 2007;204:1441–1451. doi: 10.1084/jem.20070021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pelayo R, Hirose J, Huang J, Garrett KP, Delogu A, Busslinger M, Kincade PW. Derivation of 2 categories of plasmacytoid dendritic cells in murine bone marrow. Blood. 2005;105:4407–4415. doi: 10.1182/blood-2004-07-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang GX, Lian ZX, Kikuchi K, Liu YJ, Ansari AA, Ikehara S, Gershwin ME. CD4- plasmacytoid dendritic cells (pDCs) migrate in lymph nodes by CpG inoculation and represent a potent functional subset of pDCs. J Immunol. 2005;174:3197–3203. doi: 10.4049/jimmunol.174.6.3197. [DOI] [PubMed] [Google Scholar]
- 73.Kamogawa-Schifter Y, Ohkawa J, Namiki S, Arai N, Arai K, Liu Y. Ly49Q defines 2 pDC subsets in mice. Blood. 2005;105:2787–2792. doi: 10.1182/blood-2004-09-3388. [DOI] [PubMed] [Google Scholar]
- 74.Chicha L, Jarrossay D, Manz MG. Clonal type I interferon-producing and dendritic cell precursors are contained in both human lymphoid and myeloid progenitor populations. J Exp Med. 2004;200:1519–1524. doi: 10.1084/jem.20040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ishikawa F, Niiro H, Iino T, Yoshida S, Saito N, Onohara S, Miyamoto T, Minagawa H, Fujii S, Shultz LD, Harada M, Akashi K. The developmental program of human dendritic cells is operated independently of conventional myeloid and lymphoid pathways. Blood. 2007;110:3591–3660. doi: 10.1182/blood-2007-02-071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1a-movie
Fig S1b-movie
Fig S2-movie
Fig S3