Bacterial antisense RNAs: How many are there and what are they doing? (original) (raw)
. Author manuscript; available in PMC: 2011 Jan 28.
Abstract
Antisense RNAs encoded on the DNA strand opposite another gene have the potential to form extensive base pairing interactions with the corresponding sense RNA. Unlike other smaller regulatory RNAs in bacteria, antisense RNAs range in size, from tens to thousands of nucleotides. The numbers of antisense RNAs reported for different bacteria vary extensively but hundreds have been suggested in some species. If all of these reported antisense RNAs are expressed at levels sufficient to regulate the genes encoded opposite them, antisense RNAs could significantly impact gene expression in bacteria. Here we review the evidence for these RNA regulators and describe what is known about the functions and mechanisms of action for some of these RNAs. Important considerations for future research as well as potential applications are also discussed.
Keywords: small RNA, gene regulation, transcription interference, mRNA stability
INTRODUCTION
In the past ten years there has been an explosion in the identification of small, regulatory RNAs (sRNAs) encoded on bacterial chromosomes. While some of these regulatory RNAs act by binding to and modulating protein activity, the majority of characterized sRNAs act by base pairing with target mRNAs. These base pairing sRNAs fall into two categories: _trans_-encoded and _cis_-encoded. The _trans_-encoded sRNAs are encoded at genomic locations distant from the mRNAs they regulate, and thus generally only share limited complimentarity with their targets. In part due to the ability to act via limited complimentary, many of these _trans_-encoded sRNAs have multiple mRNA targets. In a number of bacteria, this type of base pairing requires the RNA chaperone protein Hfq. Thus far the _trans_-encoded sRNAs are the most extensively characterized sRNAs and are discussed in a number of recent reviews (71, 118). In general, there has been less focus on _cis_-encoded sRNAs. These RNAs are transcribed from the DNA strand opposite another gene on bacterial chromosomes and thus have perfect complimentarity with this target. As we describe here, increasing numbers of bacterial _cis_-encoded RNAs of various sizes, which we denote antisense RNAs, are being reported and many are being characterized, raising questions about their physiological roles and mechanisms of action.
Ironically, antisense RNAs encoded on plasmids, phage and transposons were among the first regulatory sRNAs to be studied. In 1981, Tomizawa and colleagues showed that the ~108 nucleotide RNAI RNA controls the copy number of plasmid ColE1 by preventing RNAII processing to generate replication primers (105, 106). That same year, Nordström and colleagues identified the ~90 nucleotide CopA RNA, which controls the copy number of plasmid R1 by regulating the translation of the RepA replication initiator protein (99). A few years later, the 70 nucleotide RNA-OUT of the transposon Tn_10_ was found to affect transposition by repressing transposase synthesis (95). In addition, the ~ 70 nucleotide Sar RNA of bacteriophage P22 (54, 119) and the 77 nucleotide OOP RNA of bacteriophage λ (50) were reported to repress synthesis of the Ant and cII phage proteins, respectively. Another type of plasmid antisense RNA discovered early on was the ~70 nucleotide Sok RNA of plasmid R1, which represses synthesis of the toxic Hok protein responsible for postsegregational killing of cells when the R1 plasmid is lost (24). As described in several extensive reviews (14, 113, 115) much was learned about antisense RNA regulation by studies of these RNAs, along with a number of other plasmid and phage antisense RNAs, long before the chromosomally-encoded antisense RNAs described here were identified. Given the large numbers of antisense RNAs now being reported to be expressed from chromosomes, these RNAs could have a significant, as yet largely unexplored, impact on bacterial gene expression.
HOW ARE ANTISENSE RNAS BEING FOUND?
While a few chromosomally-encoded antisense RNAs were found by serendipity during the characterization of specific genes, the large increase in reported antisense RNAs has come from genome-wide searches for sRNAs and from transcriptome analysis. As illustrated by examples in Figure 1, some of the antisense RNAs are short, around 100 nucleotides in length, similar to the antisense RNAs described for plasmids and bacteriophage. However, other chromosomally-encoded antisense RNAs are much longer and in some cases correspond to the 5′ or 3′ extension of an mRNA transcribed from an adjacent protein coding gene. For example, in Listeria monocytogenes, the long 5′ untranslated region (UTR) of the mogR transcript overlaps three genes involved in flagellar synthesis encoded on the opposite strand (102). There is also significant variability in the region of overlap between the sense and antisense RNAs. Antisense RNAs can overlap the 5′-end, the 3′-end, the middle or the entire gene encoded opposite. These features have influenced how the antisense RNAs are found.
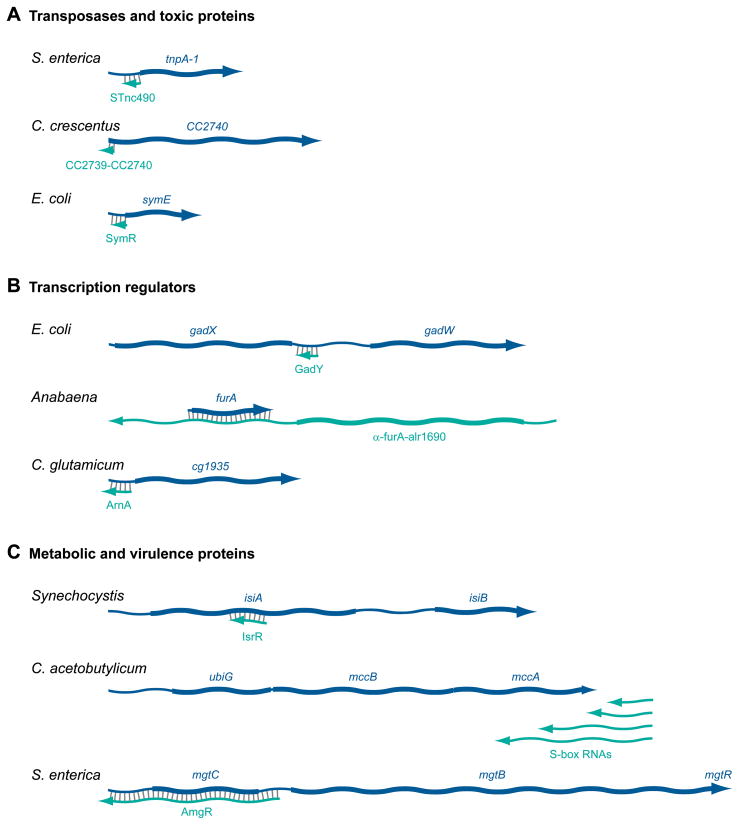
Figure 1.
Length and overlap of selected antisense RNAs with their sense transcripts. (A) RNAs antisense to transposase and toxic protein mRNAs. The ~85 nucleotide antisense RNA STnc490 of S. enterica completely overlaps the 5′-end of the IS200 transposase mRNA tnpA-1 (96), while the ~84 nucleotide CC2739-CC2740 antisense RNA of C. crescentus overlaps the IS1111A transposase mRNA CC2740 by 32 nucleotides at the 5′-end (52). In E. coli, the 77 nucleotide SymR antisense RNA completely overlaps the 5′-end of the mRNA for the SymE toxin (41). (B) RNAs antisense to mRNAs encoding transcription regulators. The 109 nucleotide GadY antisense RNA of E. coli overlaps the intergenic region of the dicistronic gadXW mRNA which encodes two transcription regulators (69, 108). The long ~2,200 nucleotide alr1690-α-furA antisense RNA of Anabaena encompasses the entire alr1690 coding region and extends through the gene encoding the FurA transcriptional regulator into its promoter and regulator regions (33). The ~131 nucleotide ArnA transcript of C. glutamicum overlaps ~ 99 nucleotides at 5′-end of the cg1935 mRNA encoding a putative transcription regulator (122). (C) RNAs antisense to genes encoding metabolic and virulence proteins. The ~177 nucleotide IsrR antisense RNA of Synechocystis overlaps the central portion of the isiA gene, which encodes a protein important under conditions of iron deficiency (13). A series of four antisense _S_-box RNAs ranging in length from 264 to 1,000 nucleotides in C acetobutylicum are transcribed from a promoter located downstream in the opposite orientation from the ubiG-mccBA operon encoding proteins important for SAM recycling. These _S_-box antisense transcripts can overlap the mccA gene by ~700 nucleotides (2). The ~1,200 nucleotide AmgR antisense RNA of S. enterica overlaps the entire first gene of the mgtCBR operon extending ~360 nucleotides into the mgtC 5′ UTR and promoter region (53). Sense RNAs are dark blue and antisense RNAs are light blue. The protein-coding regions of both classes of RNAs are indicated by thicker lines.
Prediction by computational approaches
The initial searches for chromosomally-encoded sRNAs were computational and focused on the identification of conservation and predictions of RNA structure as well as orphan promoter and Rho-independent terminator sequences in intergenic regions (reviewed in (56)). By excluding coding regions, these studies generally did not detect RNAs encoded opposite known genes. One exception was a study in which a Gapped Markov Model Index, developed based on properties of known sRNAs, tRNAs and rRNAs, was used to search both intergenic regions and regions antisense to annotated genes to predict novel sRNAs in the model organism Escherichia coli (121). This search yielded 133 candidates of which 46 candidates were predicted antisense to annotated open reading frames (ORFs). Of the five candidates tested, only one, an ~350 nucleotide RNA, was confirmed by Northern analysis. A more recent search for terminator sequences in the entire genome of the cyanobacterium Synechocystis sp. PCC6803 led to the report of 73 strong antisense RNA candidates, of which 28, ranging in size from 65 to over 1,000 nucleotides, were detected by Northern analysis and 5′-end mapping (23).
Although most of the initial computational searches for sRNAs focused on intergenic regions, subsequent studies revealed that some of the “intergenic” sRNAs are actually antisense to mRNAs. For example, in E. coli four homologous ~140 nucleotide sRNAs, now denoted Sibs (3, 85, 117), were subsequently found to be transcribed from the DNA strand opposite genes encoding the small toxic Ibs proteins (18). Additionally, the 109 nucleotide E. coli GadY sRNA (10), was shown to be encoded opposite the 3′-end of the gadX mRNA (69). Three RNAs (22–430 nucleotides in length) of the nine sRNAs initially detected in the Gram positive pathogen L. monocytogenes overlap the ends of mRNAs encoded on the opposite strand (60). Similarly, two RNAs (~60 nucleotides in length) out of six sRNAs detected in the Gram negative pathogen Helicobacter pylori are encoded opposite genes of known function (120). As a final example, in the pathogen Salmonella enteria 11 RNAs (~90–450 nucleotides in length) of the 19 sRNAs confirmed to be expressed from the intergenic regions of pathogenicity islands are partially antisense to transcripts from neighboring genes (70). In this last study, Northern and primer extension analysis of RNA taken from strains expressing the antisense RNAs on multi copy plasmids revealed that overexpression of the antisense RNAs had varied effects on the corresponding mRNA; high levels of some antisense RNAs led to decreased levels of the corresponding mRNAs, while one antisense RNA appears to undergo mutual degradation with its target mRNA and overexpression of yet another antisense RNA resulted in increased levels of the target mRNA.
As the properties of an increasing number of antisense RNAs are understood, algorithms to search for this category of sRNA will undoubtedly improve. It is worth considering the possible complication that antisense RNAs, or at least subsets of the antisense RNAs, may have features that are different from other sRNAs and thus might be hard to predict using known sRNAs as training sets. For example, antisense RNAs might not all possess Rho-independent terminators or might fall into more than one class based on their structures and functions.
Direct detection by oligonucleotide microarrays
In contrast to the computational approaches focused on intergenic regions, approaches that relied on the direct detection of sRNAs have yielded a higher proportion of antisense RNAs. In fact, antisense expression from over 75% of all annotated genes was observed in one of the first studies of genome-wide expression in E. coli using oligonucleotide microarrays with probes for the strand opposite annotated genes (88). However, no independent verification of these transcripts was carried out, and it is likely that the numbers of antisense RNAs suggested by this study are an overestimate.
Most of the initial oligonucleotide microarrays only had probes for the annotated strand of genes as well as limited coverage of both strands of intergenic regions. Nonetheless, experiments using these arrays resulted in the identification of some antisense transcripts. For example, four RNAs (~84–145 nucleotides in length) of 27 sRNAs confirmed by Northern analysis in the fresh water bacterium Caulobacter crescentus overlap the 5′- or 3′-ends of mRNAs encoded on the opposite strand (52), and 24 new antisense RNAs (100–600 nucleotides in length) were reported for the cyanobacterium Prochlorococcus (97).
The recent trend of using microarrays with oligonucleotides covering both strands of a bacterial genome has lead to the identification of many more antisense RNAs. For instance, 127 antisense transcripts ranging in size from ~200 – 3,500 nucleotides in length were described for the soil bacterium Bacillus subtilis (83). In a similar analysis of the L. monocytogenes transcriptome, seven new small antisense RNAs (77 to 294 nucleotides in length) as well as four mRNAs with long 5′ UTRs and nine mRNAs with long 3′ UTRs that overlap genes encoded on the opposite strain were identified (102).
Possible limitations to the detection of antisense RNAs by oligonucleotide microarrays include artifacts introduced by the production of the cDNAs that are generally used to probe the arrays. For example, some RNAs may be recalcitrant to reverse transcription resulting in underrepresentation of these transcripts in the final cDNA pool. For other RNAs, replication of the first strand of DNA, especially in the absence of actinomycin D, may lead to false positives (75). These problems can, at least in part, be overcome by hybridizing the RNA directly to the microarrays and detecting hybridization using labeled RNA or antibodies to RNA-DNA hybrids (36), though some RNAs may still be missed by either of these techniques. Another possible complication is cross hybridization. The oligonucleotides on many microarrays are only 25 nucleotides in length and could hybridize to homologous transcripts. A solution to this problem is the use of microarrays containing longer oligonucleotides and more stringent hybridization conditions.
Direct detection by sequencing
Several of the first chromosomally-encoded antisense RNAs were found by the sequencing of cDNA clones obtained from various pools of RNA. Such a screen in E. coli yielded the 77 nucleotide SymR and ~80 nucleotide RyjB RNAs well as cDNAs that likely correspond to longer mRNAs that overlap the 3′-end of the gene encoded opposite (42). A similar screen in the human pathogen Mycobacterium tuberculosis, led to the discovery of four antisense RNAs (75–100 nucleotides in length), which, with one exception, are encoded opposite the middle of the sense gene (4).
With the advent of improved sequencing technology, many more antisense RNAs are now being reported. For example, 89 antisense RNAs (12% of all genes) were described for the reduced-genome bacterium Mycoplasma pneumoniae (30), 96 antisense RNAs (2% of all genes) were reported for the nitrogen-fixing bacterium Sinorhyzobium meliloti (87) and 1005 antisense RNAs (22% of all genes) were reported for E. coli (12). It is important to note that the existance of the antisense RNAs was not tested by Northern analysis in any of these studies. In other experiments, 25 antisense RNAs (two of which were detected by Northern analysis) were reported for Chlamydia trachomatis, which also has a small genome (1), and 127 antisense RNAs (for which four out of nine were detected by Northern analysis) were reported for the pathogen Vibrio cholerae (55). Transcription start sites corresponding to an astonishing 969 putative antisense RNAs (46% of all genes) were reported in H. pylori (89). Northern analysis supported the expression of 21 of these RNAs (89).
These studies illustrate how antisense RNAs have now been found in a wide range of bacteria, though the percentage of annotated genes that are predicted to have antisense RNAs varies greatly. It should be noted that several of the same problems that plague antisense RNA detection by microarrays, such as the artifacts introduced by cDNA synthesis and amplification, are also problems for antisense RNA detection by sequencing.
Detection of promoter elements
Additional evidence of antisense transcription has come from the mapping of promoter elements through reporter gene fusions, computational promoter predictions or assays of the genome-wide binding of RNA polymerase or other transcription factors. In the plant bacterium Pseudomonas fluorescens, in vivo expression technology (IVET) led to the identification of 10 soil-induced transcripts (~15–1265 nucleotides in length) encoded opposite annotated ORFs (91). In E. coli, promoter elements were predicted antisense to 119 coding sequences (63) and RNA polymerase binding in chromatin immunopreciptiation assays suggested transcription antisense to 25 genes (80). However, no Northern analysis to test for the presence of the antisense RNAs was carried out. In another study, promoter activity was detected for a number of chromosomal lacZ fusions antisense to annotated genes in E. coli, but no transcripts were detected for the eight putative antisense RNAs examined by Northern analysis. This finding suggests that although the chromosome contains numerous promoter sequences, the transcripts may never accumulate (43). As more promoters, transcription start sites and RNA polymerase binding sites are mapped, it will be useful to determine whether these different approaches predict overlapping sets of antisense RNAs.
Considerations
The prospect of so many antisense RNA regulators is exciting, but some caution is warranted. The expression of only a few of the antisense transcripts has been confirmed by independent means, which is critical given the possible artifacts mentioned above. In addition, some of the Northern blots used to verify antisense RNAs show ambiguous bands. Finally, an even smaller number of antisense RNAs have an identified function. It is possible that some of the transcripts are a result of nonspecific transcription or occasional readthrough from upstream or downstream genes and thus do not have physiological functions. In addition, it is worth considering whether both the sense and antisense RNAs are expressed in the same cell. Recent genome-wide quantitation of transcript levels suggests that many transcripts are present at levels such that there is less than one RNA molecule per cell (74). Thus although both sense and antisense transcripts are detected in the pools of cells used for the experiments, they may actually never be present in the same cell. Finally, it is conceivable that a subset of antisense RNAs with true physiological functions are so short-lived that they can never be detected.
Further confirmation and characterization of the antisense RNAs is critical before strong conclusions about the total number of antisense RNAs can be drawn. For these studies it is necessary to eliminate the expression of the antisense RNA to ensure that the observed signal is due to the RNA and to determine whether there are any phenotypes associated with the lack of the RNA. This may require the introduction of mutations that eliminate the antisense RNA promoter, but are neutral with respect to the sense gene, or the synthesis of an anti-antisense transcript to neutralize the antisense RNA. Many studies are carried out with overexpression of the antisense RNA. This approach can provide important insights yet also introduce artifactual effects that are not observed when the antisense RNA is expressed at endogenous levels. Evidence for regulated expression as well as conservation in closely related species will give weight to the conclusion that a specific antisense RNAs has a physiological role.
WHAT ARE ANTISENSE RNAS DOING?
Antisense transcripts have been reported opposite genes encoding a wide variety of activities, but very few of these antisense RNAs have been examined for function. However, for those antisense RNAs for which an effect on sense gene expression has been shown, the proteins encoded by the sense genes can be grouped into some general categories, providing the first functional insights.
Repression of transposase and toxic protein synthesis
Several antisense RNAs are encoded opposite genes encoding transposases (Figure 1A). One of the first antisense RNAs to be discovered, RNA-OUT of the transposon Tn_10_, was found to repress transposition by reducing transposase levels. Two antisense RNAs discovered in S. enterica (70, 96), two in C. crescentus (52) and three in L. monocytogenes (102) also are encoded opposite transposase genes. Thus an important function of antisense RNAs in bacteria, as in eukaryotes (reviewed in (59)), could be to inhibit transposition.
An increasing number of chromosomally-encoded antisense RNAs that down regulate the synthesis of potentially toxic proteins, as was first found for the Sok RNA of plasmid R1, are also being discovered ((19), reviewed in (17) and (26)) (Figure 1A). Most of the proteins whose synthesis is repressed by these antisense RNAs are small (< 50 amino acids), hydrophobic and toxic at high levels. Although the cellular roles of the proteins are not yet known, it is clear that the synthesis of certain potentially toxic proteins is tightly repressed. For example, the E. coli SymE protein is maintained at low levels by the LexA repressor of the SOS response, the SymR antisense RNA and the Lon protease (41).
The expression of only a few antisense RNAs has been assayed under multiple conditions. However some of the RNAs antisense to transposase and toxin genes, such as SymR, appear to be present constitutively in the cell. All of these antisense RNAs have been found to act as repressors, consistent with a role in the continuous tight repression of potentially detrimental proteins.
Regulating the levels of transcription regulators
Other antisense RNAs have been shown to regulate the synthesis of transcription regulators. In E. coli the GadY RNA was originally identified in a computational search of intergenic regions (10), but was later found to overlap the 3′ UTR of the gadX mRNA (69) (Figure 1B). Both gadX and the downstream gadW gene encode transcription regulators of the acid stress response, and further studies of the transcripts in the gadXW region showed that the two genes are co-transcribed (108). Overexpression of the GadY RNA results in processing of the gadXW mRNA giving rise to separate gadX and gadW transcripts that accumulate to higher levels than the full length mRNA (69, 108). Removal of the region of _gadX_-GadY base pairing eliminates processing and leads to decreased levels of the gadX mRNA, even with GadY overexpression (69). GadY is specifically induced in stationary phase in an σS dependent manner, and the increased levels of the GadX transcription regulator that result from the GadY-directed processing lead to increased expression of GadX target genes (69).
In many bacteria, Fur (ferric uptake regulator) is a key transcription repressor of genes involved in iron uptake and metabolism in the presence of high iron levels. In the cyanobacterium Anabaena sp. PCC 7120, a 2,200 nucleotide antisense transcript, detected by Northern analysis carried out with a double-stranded probe, was shown to decrease furA expression and translation (33). End mapping revealed that the antisense transcript starts near the promoter of alr1690 (encoded downstream and on the strand opposite furA) and overlaps the entire furA mRNA as well as the furA promoter (Figure 1B). Elimination of the alr1690-α-furA antisense transcript resulted in increased levels of the FurA protein (33) as well as the FurA-regulated alr3808 transcript implicating alr1690-α-furA in FurA regulation (34). Promoters of both furA and the alr1690-α-furA appear to be bound by NtcA, a regulator of nitrogen metabolism genes (57). A number of proteins involved in nitrogen fixation have iron cofactors, explaining the need to co-regulate iron acquisition and nitrogen metabolism. However, it is not clear how the furA and the alr1690-α-furA transcripts are regulated relative to each other. It is intriguing that regulatory RNAs, in the form of _trans_-encoded sRNAs in many bacteria and iron response elements in the 5′ and 3′ UTRs of a number of eukaryotic mRNAs, are components of the iron response in many different organisms (reviewed in (61, 84).
A promoter screen in the Gram-positive bacterium Corynebacterium glutamicum led to the identification of the ~130 nucleotide antisense RNA, ArnA, encoded opposite the 5′-end of the cg1935 gene (122) (Figure 1B). This gene encodes a protein that shares similarity with the GntR family of transcription regulators. The ArnA RNA was only detected after cells were incubated at high temperature suggesting possible regulation by the heat shock sigma factor SigH. The effects of ArnA on cg1935 were examined using a reporter construct, which gave reduced expression at high temperature in the absence of ArnA suggesting a potential role for ArnA in stabilizing the mRNA encoding the putative transcription regulator. However, further experiments are needed to clarify the mechanism of action and physiological role of ArnA. The L. monocytogenes RliH RNA also overlaps a gene encoding a putative transcription regulator (60), and we predict that still other antisense RNAs will be found to modulate the levels of transcription regulators, either positively (like GadY or ArnA) or negatively (like alr1690-α-furA).
Modulating the levels of metabolic and virulence proteins
Antisense RNAs have also been shown to regulate the expression of some metabolic enzymes and virulence factors. One example is the 177 nucleotide IsrR antisense RNA encoded opposite the iron-stress induced IsiA protein in the cyanobacterium Synechocystis sp PCC6803 (13) (Figure 1C). The levels of IsiA, a protein that enhances light absorption by forming an antenna ring around photosystem I, are elevated under conditions of iron deficiency and reduced under conditions of iron excess. Studies of IsiA expression revealed that the response to iron is controlled, at least in part, by the IsrR RNA, which was first detected by Northern analysis of the transcripts in the isiAB region. IsrR overexpression leads to decreased levels of IsiA, and depletion of IsrR leads to premature isiA expression.
In Clostridium acetobutylicum, computational studies predicted an antisense RNA containing a _S_-adenosyl methionine (SAM)-responsive, _S_-box riboswitch element convergent to the ubiG-mccBA operon, which encodes enzymes involved in the conversion of SAM to cysteine (86). Four _S_-box containing antisense RNAs of 264–1000 nucleotides in length subsequently were detected by Northern analysis (Figure 1C), and a reduction in MccB protein activity in the presence of sulfur sources was shown to be dependent on the presence of the promoter for the _S_-box antisense RNAs (2). Since the longer antisense RNAs were not detected in the presence of methionine, this led to the model that, in the absence of SAM, there is readthrough of the _S_-box riboswitch generating the antisense RNAs, which in turn leads to a reduction of MccB synthesis.
Northern analysis using double-stranded probes also led to the detection of the 1,200 nucleotide AmgR RNA encoded opposite the S. enterica mgtCBR operon associated with virulence and survival in macrophages (53). AmgR is transcribed from within the mgtC-mgtB intergenic region and overlaps the entire mgtC gene extending into the mgtC 5′ UTR (Figure 1C). Expression of both the antisense AmgR RNA and the sense mgtCBR mRNA was found to be induced by the response regulator of the PhoQ/PhoP two component system in response to low Mg2+ levels. The presence of AmgR is associated with degradation of the mgtC mRNA and a corresponding decrease in MgtC, and to a lesser extent MgtB, protein levels. MgtC protein levels in turn impact S. enterica virulence in mice. A S. enterica strain overexpressing AmgR was less virulent while an amgR promoter mutant strain was more virulent in mice compared to the wild type control strain.
IsrR, the _S_-box RNAs and AmgR illustrate how antisense RNAs might be a factor in modulating the levels of a wide range of metabolic and virulence proteins. While not well characterized, antisense RNAs also are proposed to impact flagellar synthesis in H. pylori (120), L. monocytogenes (102) and S. enterica (116). In all of these examples, antisense RNAs introduce an additional layer of regulation to systems whose expression is already extensively regulated.
Reasons for antisense regulation
One relevant question is why regulation by an antisense RNA might be advantageous over other types of regulation. From looking at the examples described above two possible answers arise. First, antisense RNAs could provide an advantage when the levels of a particular protein need to be repressed very tightly and expressed under very select circumstances, as in the case of a transposase or toxin. Second, many of the characterized antisense RNA targets are subject to extensive regulation, where the antisense RNAs provide yet one more level of control. For example, the levels of S. enterica MgtC are regulated at the transcriptional level by PhoPQ, at the posttranscriptional level by the AmgR antisense RNA and at the level of protein stability by the MgtR peptide (53). It is interesting to note that for the mgtC mRNA-AmgR RNA pair, the same regulator controls expression of both transcripts. The same is also true for other sense-antisense pairs. While this arrangement at first seems counterintuitive, it may allow for the establishment of desired regulatory circuits.
So far, antisense RNAs appear to fall into two classes with respect to how their transcription is regulated; some appear to be expressed constitutively like the E. coli SymR RNA, while for others, transcription is tightly controlled. Two additional examples of antisense RNAs that show strong regulation can be found in B. subtilis. Here, expression of a 750 nucleotide RNA encoded opposite the 3′-end of the yabE gene is regulated by the extracytoplasmic σ factors, σX and σM (15), while expression of the 280 nucleotide SurA RNA encoded opposite the 5′-end of yndL is under the indirect control of Spo0A, the master regulator of sporulation (94). Other trends in the expression and function of antisense RNAs should become obvious as more RNAs are characterized. For example, it will be interesting to see how many short and long antisense RNAs have physiological functions and whether the short antisense RNAs have functions that are different from the longer antisense RNAs.
HOW DO ANTISENSE RNAS ACT?
As already mentioned, antisense RNAs can overlap the 5′-end, the 3′-end, the middle, or the entire gene encoded opposite. It is also possible that while base pairing may take place at one position, the effects of the base pairing may be propagated to another region of the sense RNA. The sense and antisense RNAs also show a range of expression patterns. In some cases, both transcripts show similar patterns of expression; both high or both low under a specific condition. In other cases the transcripts show opposing patterns of expression, with the level of one RNA high and the other low under a specific condition. All of these features affect how the antisense RNA might impact the transcription, stability or translation of the sense RNA.
Transcription interference and attenuation
Antisense RNAs can alter transcription of the genes encoded on the opposite strand in two different ways; transcription interference or transcription attenuation (Figure 2A and 2B). Transcription interference occurs when transcription from one promoter is suppressed by a second promoter present in cis (reviewed in (90)). This effect was studied for two convergent bacteriophage 186 promoters that produce transcripts overlapping by 62 base pairs at their 5′-ends (7). In this study, a stronger promoter reduced the activity of the weaker promoter by 5.6 fold. When the promoters were oriented divergently, but the transcripts still maintained their regions of complimentarity, transcription of the weaker promoter was not affected by transcription from the stronger promoter. In addition, the introduction of a terminator before the convergent weak promoter resulted in reduced interference. These results led to the conclusion that the convergent orientation of the promoters was the source of the interference rather than base pairing.
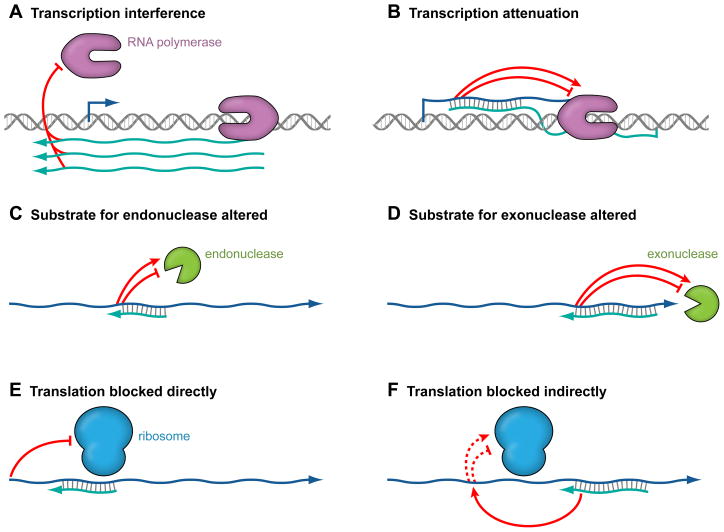
Figure 2.
Mechanisms by which antisense RNAs act. Antisense RNAs can induce transcription interference (A), where transcription from one promoter blocks transcription from a second promoter by preventing RNA polymerase from either binding or extending a transcript encoded on the opposite strand. Transcription interference does not involve basepairing and does not occur when the antisense RNA is provided in trans. In transcription attenuation (B), base pairing of the antisense RNA to the target RNA causes changes in the target RNA structure ultimately affecting transcription termination. Antisense RNAs can also affect target RNA degradation by endonucleases (C) and exonucleases (D). In these cases, base pairing between the sense and antisense RNAs can directly either generate or block a ribonuclease target site. Antisense RNAs can also indirectly affect the binding of the ribonuclease at a distance from the site of base pairing. Finally, antisense RNAs can directly block ribosome binding (E) or indirectly positively or negatively impact ribosome binding by affecting the target mRNA structure (F). The sense RNAs are indicated in dark blue while the antisense RNA are shown in light blue.
Transcription interference has been proposed to be the mechanism by which the _S_-box antisense RNAs of C. acetobutylicum block expression of the ubiG-mccBA mRNA (2). The antisense RNAs generated from the promoter of the _S_-box RNA can overlap the ubiG-mccBA mRNA by up to 700 base pairs, but the levels of the ubiG-mccBA mRNA were unchanged in a number of ribonuclease mutants suggesting the antisense RNAs do not impact mRNA processing. Based on this observation and the finding that the antisense RNA provided in trans did not affect ubiG-mccBA mRNA levels, transcription interference was inferred, but this has not been tested directly.
Antisense RNAs also can alter target mRNA transcription by inducing attenuation whereby transcription of the opposite strand is prematurely terminated. In a few cases, generally found on plasmids, base pairing of the antisense RNA to the mRNA has been shown to induce the formation of a terminator structure in the target mRNA. For example, the 427 nucleotide RNAβ, encoded opposite the fatDCBA-angRT iron transport-biosynthesis operon on the virulence plasmid pJM1 in the fish pathogen Vibrio anguillarum, was reported to be required for transcription attenuation (98). In vitro transcription and S1 nuclease protection assays indicated that the fatDCBA-angRT transcript is terminated after interaction of a stem-loop in the fatA-angR intergenic region, with stem-loops of RNAβ. Transcription termination results in increased levels of the fatA portion of the mRNA compared to the downstream angRT portion providing a mechanism for discoordinate expression within an operon. How frequently transcription interference or attenuation mechanisms involve antisense RNAs remains to be determined.
RNA cleavage
Antisense RNAs can also impact the stability of a target RNA by either promoting or blocking cleavage by endoribonucleases or exoribonucleases (Figure 2C and 2D). In many bacteria, two major endoribonucleases have been linked to antisense RNA-induced target mRNA cleavage. The first, RNase III, cleaves double-stranded RNA and is generally associated with rRNA processing (reviewed in (8)). Its ability to specifically degrade double stranded RNA complexes makes it an ideal candidate for processing antisense RNAs base paired with their targets. In fact, RNase III was found to be responsible for cleavage of the copT-copA and hok-sok plasmid-based sense-antisense RNA pairs in early studies (5, 25). Similarly, RNase III was shown to cleave two sRNAs RyeA (also called SraC) and RyeB encoded opposite each other in E. coli (112). The antisense RNA-directed cleavage could generate two target RNA halves that are both less stable, both more stable, or differentially stabilized relative to the original target transcript. The second endonuclease reported to be involved in antisense RNA directed processing is RNase E, which cleaves single-stranded RNA. RNase E is a component of the degradasome, a multi-protein complex that globally affects mRNA stability, and has been shown to interact with Hfq (reviewed in (8)). The AmgR RNA of S. enterica was reported to induce degradation of the mgtC mRNA in a manner that requires RNase E but not RNase III (53). Exactly how a double stranded region of RNA might stimulate RNase E activity is not yet clear, though it is conceivable that the 5′-end of an antisense RNA could provide a monophosphate which has been shown to stimulate RNase E activity (9). It is also possible that base pairing might block an RNase E recognition site thus leading to increased stability of the target RNA.
RNase III and RNase E likely are not the only endonucleases involved in antisense RNA -directed processing. While RNase III was responsible for some of the OOP RNA-directed cleavage of the _c_II-O mRNA of bacteriophage λ, processing was still observed within the regions of base pairing, albeit with a slightly different pattern, in a strain lacking RNase III (51). However, determining what endonucleases are responsible for a particular cleavage event can be difficult due to redundancies between cellular RNases. The fact that several of these enzymes are essential and show autoregulation and crossregulation also complicates this analysis.
In addition to providing substrates for endonucleases, the base pairing between a sense and an antisense RNA might impact the ability of an exonuclease to degrade a particular target RNA. For example, the region of base pairing might block an exoribonuclease similar to how stem-loops within a transcript can block digestion. Additionally, the base pairing could alter the target RNA structure so that the ends are more or less susceptible to cleavage. Since only a few studies have been carried out to examine the roles of exonucleases in sRNA and antisense RNA-directed processing, this is an important direction for future studies.
Translation block
Many _trans_-encoded sRNAs base pair with the Shine Dalgarno sequences of their target mRNAs thereby preventing ribosome binding and protein translation (reviewed in (71, 118)). Antisense RNAs for which complimentarity extends into the 5′ untranslated region of the mRNA target may act by a similar mechanism (Figure 2E). The SymR antisense RNA of E. coli overlaps the 5′-end of the symE mRNA (41). In a strain with a symR promoter mutation, the levels of the symE mRNA were increased ~3-fold while the levels of the SymE protein were increased ~10-fold suggesting some of the repression by SymR occurs by blocking translation.
Similarly, the alr1690-α-furA antisense RNA overlaps the length of the furA transcript including the Shine Dalgarno sequence. Elimination of the alr1690-α-furA transcript resulted in increased levels of the FurA protein consistent with a possible block in translation (33), but further experiments are needed to verify this. Ribosome binding and mRNA stability are often linked, presenting a complication in evaluating whether the primary effect of sense-antisense pairing is a block in translation. For example, the AmgR RNA overlaps the Shine-Dalgarno sequence of its target mRNA mgtC, yet has been proposed to destabilize the mgtC mRNA in an RNase E dependent manner (53). Base pairing between the sense and antisense RNA also could positively or negatively impact translation by altering the structure of the target RNA at a ribosome binding site distant from the region of base pairing (Figure 2F).
Dual functions
Modulating the transcription, stability or translation of a _cis_-encoded gene might not be the only function of a particular antisense RNA. One of the short antisense RNAs identified in P. fluorescens was reported to encode a small protein (92), and some of the longer antisense RNAs such as alr1690-α-furA are known to encode proteins. Subsets of these RNAs may have dual roles as mRNAs and base pairing RNAs though others might function solely as mRNAs. Antisense RNAs also might act as both _cis_- and _trans_-encoded base pairing RNAs. Among the shorter antisense RNAs, the E. coli GadY RNA has been shown to bind to Hfq, the RNA chaperone required by _trans_-encoded base pairing sRNAs but thus far not thought to be required by _cis_-encoded base pairing RNAs. This suggests GadY might base pair with other mRNA targets (69). Similarly, the RliE RNA of L. monocytogenes (60) and the ASdes and ASpks RNAs of M. tuberculosis (4) have been predicted to base pair with a number of targets other than the transcript encoded opposite the antisense RNA gene. Further investigation is required to determine whether many antisense RNAs have dual roles.
HOW DO ANTISENSE RNAS BASE PAIR?
The mechanisms of base pairing between an antisense RNA and its target have been studied most extensively for the RNAs found encoded on plasmids and transposons using in vitro structure probing and mobility shift experiments. Two general mechanisms for how base pairing proceeds have been proposed. In the first mechanism, denoted a one-step mechanism, an initial point of contact between the antisense RNA and the mRNA target leads to complete duplex formation. In the second mechanism, denoted a multi-step mechanism, an initial transient interaction between the antisense RNA and the mRNA target requires stabilization by a protein or the formation of additional base pairs. The stable complex then can go on to form a complete duplex between the antisense RNA and the mRNA target, though this proceeds slowly and, in cases where it has been examined, does not appear to be required for regulation. In many cases of base pairing, stem loops containing a YUNR U-turn motif (a motif containing a pyrimidine (Y) followed by a uracil (U), any nucleotide (N), and a purine) are important for the RNA-RNA interactions (reviewed in (20)).
One-step mechanism
The base pairing between the Hok and Sok RNAs of plasmid R1 proceeds via a one-step binding mechanism whereby a single-stranded 5′ region of the antisense Sok RNA base pairs with a stem loop containing a YUNR U-turn motif in a processed version of the hok mRNA (21, 22, 100). Initial base pairing then progresses to full duplex formation between the Sok RNA and the hok mRNA preventing Hok protein translation. The base pairing between the transposase mRNA and RNA-OUT of the transposon Tn_10_ similarly involves one-step base pairing. The YUNR U-turn loop of RNA-OUT and the 5′ region of the transposase mRNA first interact since mutations in either of these regions reduced base pairing (46). Again, a full duplex is subsequently formed.
Multi-step mechanism
Base pairing between RNA II and RNAI of plasmid ColE1 and the repA mRNA and CopA of plasmid R1 proceeds via a multi-step mechanism. The first step of base pairing between RNAI and RNAII is the formation of a transient “kissing complex” through the single stranded regions of RNAI and RNAII stem loops which both contain YUNR motifs (103). In the second step, the _R_NA _o_ne _m_odulator protein, Rom (also named Rop) recognizes the structure of the “kissing complex” (rather then the nucleotide sequence) and stabilizes this RNAI-RNAII complex (81, 104, 107). The stabilized interaction between the loop structures then brings the 5′-end of RNAI in close proximity to its complimentary region in the RNAII transcript, facilitating complete duplex formation (103, 104).
The first step in the interaction between the CopA antisense RNA and the CopT target region in the repA mRNA leader sequence also involves the formation of a “kissing complex”. This requires stem loop II of CopA contacting the YUNR motif in the CopT stem loop II (22, 35, 48, 49, 77–79). The formation of an extended “kissing complex” involving nucleotides other then the initial stem loop II region is required to stabilize the initial R1 “kissing complex”. After formation of the extended “kissing complex”, the base pairing progresses to a structure in which two helices are formed between CopA and CopT. This is further stabilized by a third helix generated by base pairing between a stretch of single stranded nucleotides in the 5′-end of CopA with a complimentary region in CopT (47, 48, 58). This stabilized structure is sufficient for replication control as formation of a completely base paired duplex is very slow in vitro and is unnecessary for replication control in vivo (58, 78, 114).
While the sense-antisense base pairing interactions for the transposon and plasmid RNAs have been characterized extensively, much less is known about chromosomal sense-antisense base pairing. However, a recent study of the interactions between the chromosomally-encoded ibs mRNAs and Sib antisense RNAs suggest that base pairing may proceed by similar mechanisms (31). Structure probing of the interactions between the Sib and ibs RNAs revealed that base pairing is initiated via two regions. A YUNR U-turn motif, located in one target recognition domain (TRD1) of the SibC RNA recognizes a complimentary YUNR-U turn motif in the ibsC mRNA. A second domain (TRD2) within a single stranded region between the adjacent stems of SibC could also initiate base pairing with the ibsC mRNA. The interactions between both of these target regions led to more complete complex formation but did not result in formation of a fully base paired duplex. Mutant versions of SibC showed that either TRD1 (opposite the ibsC ORF) or TRD2 (opposite the ibsC translation initiation region) were able to repress IbsC protein synthesis independently, perhaps through different mechanisms, indicating complete duplex formation may not be required to prevent IbsC toxicity. While the TRD1-mediated interaction involved a YUNR U-turn motif, the TRD2 region is similar to the single stranded regions of Sok and RNA-OUT that are involved in pairing. Whether other chromosomally encoded antisense RNAs use the loop-loop YUNR interactions, a loop-single strand interaction, a combination of both, or completely different base paring interactions remains to be determined. It will also be of interest to test whether the effects are reversible, since it is conceivable that RNA helicases may dissociate base paired RNAs.
Additional factors, which are likely to influence antisense RNA-target RNA base pairing and have not been examined in detail, include the relative amounts of the two transcripts as well as other proteins interacting with the transcripts. The levels of both the sense and antisense RNAs have been determined in very few cases. For the _symE_-SymR pair in E. coli, the levels of the SymR antisense RNA are in significant excess (10-fold higher) over the symE mRNA (41). In addition, the structures of the RNAs and the availability of different regions for base pairing may vary, as the RNAs are being transcribed or processed. Finally, portions of one or both of the RNAs may be translated and bound by ribosomes which undoubtedly will also impact the ability of the RNAs to base pair.
CAN ANTISENSE RNAS BE EXPLOITED?
Not surprisingly, the use of synthetic antisense RNAs as antibiotics and in biotechnology has been under investigation for quite some time. Two general approaches for delivering synthetic antisense transcripts have been pursued: expression of an antisense transcript from a gene introduced into the cell and the direct delivery of antisense oligonucleotides. Degradation is a problem with both in vivo synthesized RNAs and the direct delivery of oligonucleotides. To circumvent this problem, antisense RNAs that form more stable hairpin structures using paired termini have been used (66). In addition, antisense oligonucleotides have been modified by adding polyamide backbones (PNAs), substituting morpholine rings for the deoxyribose rings (PMOs), and replacing the internucleoside linkages with phosphorothioates (PS-ODNs) (reviewed in (82)). However, the cell walls of bacteria can inhibit uptake of antisense oligonucleotides, PNAs and PMOs, limiting their applicability. To address this problem, cell permeable cationic peptides have been conjugated to PNAs and PMOs increasing cellular uptake and potency and extending retention of the peptide-PNA within the cell (28, 68)
Synthetic antisense RNAs have already been used in a number of applications. They have been shown to inhibit growth of E. coli (28, 62, 68, 101), S. enterica (64), Staphylococcus aureus (67) and M. tuberculosis (32) when targeted to essential genes. Since antisense RNAs allow for the conditional repression of target genes, they also have been employed to study bacterial growth and metabolism (6, 27, 40, 76, 110) and to characterize known or putative virulence factors (38, 65). Antisense RNAs have been used to sensitize other bacteria to antibiotics (37, 39, 44), identify novel antibiotics (73, 123), identify antibiotic targets (29, 32, 39), or clarify the mechanisms of action of potential new drugs (39). In addition, antisense technology has allowed the induction of “bacterial suicide” without the need for antimicrobial compounds. For example, in E. coli introduction of a PNA complementary to the Sok antisense RNA resulted in the synthesis of the toxic Hok protein and cell death (16). Finally, antisense RNA technology has been used to alter bacterial gene expression in industrial processes in order to produce chemicals more efficiently with fewer unwanted byproducts (11, 93, 109, 111) and to optimize protein production (45). C. acetobutylicum, an important industrial bacteria, is able to ferment a number of complex carbohydrate sources, like cellulose, into important industrial solvents, acids, and biofuels such as ethanol, acetone and butanol (reviewed in (72)). Genetic manipulations of Clostridium are difficult, but antisense technology has been successful in modifying C. acetobutylicum primary metabolism to produce the desired industrial compounds and biofuels (11, 93, 109, 111).
Despite the success of antisense RNA technology, a number of issues still need to be addressed. In general synthetic antisense RNAs are targeted to the Shine-Dalgarno sequence, however different regions of specific target mRNAs may correlate with more efficient repression. Additionally, antisense oligonucleotides, due to their short length, can have “off target” effects. The use of multiple oligonucleotides against the target RNA, which should not all have the same off target effects, can be a control for this problem. Undoubtedly, a greater understanding of endogenous target RNA-antisense RNA base pairing and target inhibition will facilitate the engineering of better synthetic antisense RNAs for clinical, industrial, or laboratory use.
PERSPECTIVES
The study of antisense RNAs is one of the most exciting areas of regulatory RNA research in bacteria given the many questions that remain to be answered. With the recent advances in transcriptome analyses, hundreds of antisense RNAs are being reported. How many of these newly reported antisense RNAs have physiological functions? Will most have dual functions as mRNAs and base pairing RNAs or as basepairing RNAs that act on both _cis_- and _trans_-encoded targets? Many of the characterized antisense RNAs repress transposon and toxic protein synthesis and modulate the expression of genes encoding transcription regulators or other genes that show complex regulation. Will the targets of newly identified functional antisense RNAs fall into these same categories? Known antisense RNAs can act through a number of mechanisms including transcription interference and attenuation, RNA stability or ribosome binding. Will one or another mechanism predominate as more RNAs are characterized? The length of antisense RNAs can vary from tens to a few thousand nucleotides. Will antisense RNAs of different lengths be associated with different mechanisms of action? Studies of short antisense RNAs have revealed that base pairing with the target RNA generally is mediated by loop-loop interactions. Will longer antisense RNAs base pair using the same mechanisms? With more and more tools available for the identification of chromosomally-encoded antisense RNAs and increasing interest in antisense RNAs, answers to these questions should be forthcoming.
SUMMARY POINTS.
- Bacterial chromosomes encode a number of small, regulatory RNAs, many of which act by base pairing. Base pairing RNAs fall in to categories. _Trans_-encoded sRNAs are encoded at genomic locations distinct from their target mRNA(s) and act via limited base pairing. _Cis_-encoded, or antisense RNAs, are encoded on the DNA strand opposite other genes and thus can act via more extensive base pairing.
- With recent advances in direct detection methods, the numbers of antisense RNAs reported has increased dramatically from just a few to hundreds. However, the validation and functional characterization for most of these newly reported antisense RNAs is lacking.
- The first antisense RNAs to be discovered (associated with plasmids, phage, and transposons) were small, generally only a few hundred nucleotides in length. However, several of the newly-described chromosomally-encoded antisense RNAs are reported to be larger then 1000 nucleotides in length, in some cases overlapping entire genes and themselves encoding proteins.
- Antisense RNAs have been shown to repress target mRNAs encoding proteins, such as transposons and toxic proteins, that have the potential to be detrimental to the cell. They also have been shown to positively and negatively impact the expression of transcription regulators as well as a number of other metabolic and virulence proteins, many of which are regulated extensively at other levels.
- Antisense RNAs can act by altering their target gene transcription through interference or transcription attenuation. Antisense RNAs can also provide or block recognition sites for endonucleases as well as exonucleases. In addition, some antisense RNAs can act like _trans_-encoded sRNAs to block ribosome binding and translation. Finally, antisense RNAs can have dual functions and act as both mRNAs and antisense RNAs or both _cis_- and _trans_-encoded RNAs.
- Antisense RNAs can base pair via stem loops containing YUNR U-turn motifs or via single stranded regions of sequences complimentary to regions on their target mRNAs. Base pairing can proceed by either a one-step mechanism whereby an initial contact proceeds to duplex formation or by a two-step mechanism whereby the initital “kissing complex” is first stabilized by a protein or additional contacts before proceeding to duplex formation.
FUTURE ISSUES.
- The hundreds of antisense RNAs now being reported need to be further validated and functionally characterized before generalizations regarding the extent of antisense transcription in bacterial genomes can be made.
- Future studies should clarify whether antisense RNAs fill a specific regulatory niche in the cell.
- Much remains to be learned about the mechanisms of antisense RNA action and base pairing particularly for longer antisense RNAs.
Acknowledgments
We thank Susan Gottesman, Jörg Vogel and members of the Storz lab for comments regarding the review.
ABBREVIATIONS/ACRONYMS
IVET
in vivo expression technology
ORF
open reading frame
SAM
_S_-adenosyl methionine
sRNA
small regulatory RNA
TRD
target recognition domain
UTR
untranslated region
YUNR
a motif containing a pyrimidine (Y) followed by a uracil (U), any nucleotide (N), and a purine
KEY TERMS/DEFINITIONS
_cis_-encoded RNA
regulatory RNA encoded on the DNA strand opposite its target mRNA; also termed antisense RNA
Hfq
RNA chaperone protein required for base pairing by many _trans_-encoded sRNAs
target mRNA
RNA regulated by base pairing with a _cis_- or _trans_-encoded RNA
kissing complex
initial base pairing interaction between an antisense RNA and its mRNA target
RNase III
endoribonuclease that cleaves double stranded RNA; generally involved in ribosomal RNA processing
RNase E
endoribonuclease that cleaves single stranded RNA; part of the degradasome
_trans_-encoded sRNA
small, regulatory RNA encoded at a genomic location distinct from the target RNA
Contributor Information
Maureen Kiley Thomason, Email: kileyma@mail.nih.gov.
Gisela Storz, Email: storz@helix.nih.gov.
LITERATURE CITED
- 1.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acid Res. 2010;38:868–77. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.André G, Even S, Putzer H, Burguière P, Croux C, et al. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acid Res. 2008;36:5955–69. doi: 10.1093/nar/gkn601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EGH, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Microbiol. 2001;11:941–50. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 4.Arnvig KB, Young DB. Identification of small RNAs in Mycobacterium tuberculosis. Mol Microbiol. 2009;73:397–408. doi: 10.1111/j.1365-2958.2009.06777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomberg P, Wagner EGH, Nordström K. Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 1990;9:2331–40. doi: 10.1002/j.1460-2075.1990.tb07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borden JR, Jones SW, Indurthi D, Chen Y, Papoutsakis ET. A genomic-library based discovery of a novel, possibly synthetic, acid-tolerance mechanism in Clostridium acetobutylicum involving non-coding RNAs and ribosomal RNA processing. Metab Eng. 2010;12:268–81. doi: 10.1016/j.ymben.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell. 2004;14:647–56. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Carpousis AJ, Luisi BF, McDowall KJ. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- 9.Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, et al. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems. 2002;65:157–77. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 11.Desai RP, Papoutsakis ET. Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum. Appl Environ Microbiol. 1999;65:936–45. doi: 10.1128/aem.65.3.936-945.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dornenburg JE, DeVita AM, Palumbo MJ, Wade JT. Widespread antisense transcription in Escherichia coli. MBio. 2010 doi: 10.1128/mBio.00024-10. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dühring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci USA. 2006;103:7054–8. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eguchi Y, Itoh T, JT Antisense RNA. Annu Rev Biochem. 1991;60:631–52. doi: 10.1146/annurev.bi.60.070191.003215. [DOI] [PubMed] [Google Scholar]
- 15.Eiamphungporn W, Helmann JD. Extracytoplasmic function sigma factors regulate expression of the Bacillus subtilis yabE gene via a cis-acting antisense RNA. J Bacteriol. 2009;191:1101–5. doi: 10.1128/JB.01530-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faridani OR, Nikravesh A, Pandey DP, Gerdes K, Good L. Competitive inhibition of natural antisense Sok-RNA interactions activates Hok-mediated cell killing in Escherichia coli. Nucleic Acid Res. 2006;34:5915–22. doi: 10.1093/nar/gkl750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev. 2008;72:579–89. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, et al. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol Microbiol. 2008;70:1076–93. doi: 10.1111/j.1365-2958.2008.06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38:3743–59. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franch T, Gerdes K. U-turns and regulatory RNAs. Curr Opin Microbiol. 2000;3:159–64. doi: 10.1016/s1369-5274(00)00069-2. [DOI] [PubMed] [Google Scholar]
- 21.Franch T, Gultyaev AP, Gerdes K. Programmed cell death by hok/sok of plasmid R1: processing at the hok mRNA 3′-end triggers structural rearrangements that allow translation and antisense RNA binding. J Mol Biol. 1997;273:38–51. doi: 10.1006/jmbi.1997.1294. [DOI] [PubMed] [Google Scholar]
- 22.Franch T, Petersen M, Wagner EGH, Jacobsen JP, Gerdes K. Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J Mol Biol. 1999;294:1115–25. doi: 10.1006/jmbi.1999.3306. [DOI] [PubMed] [Google Scholar]
- 23.Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerdes K, Bech FW, Jørgensen ST, Løbner-Olesen A, Rasmussen PB, et al. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986;5:2023–9. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerdes K, Nielsen A, Thorsted P, Wagner EGH. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable Hok, SrnB and PndA effector messenger RNAs. J Mol Biol. 1992;226:637–49. doi: 10.1016/0022-2836(92)90621-p. [DOI] [PubMed] [Google Scholar]
- 26.Gerdes K, Wagner EGH. RNA antitoxins. Curr Opin Microbiol. 2007;10:117–24. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Goh S, Boberek JM, Nakashima N, Stach J, Good L. Concurrent growth rate and transcript analyses reveal essential gene stringency in Escherichia coli. PLoS One. 2009;4:e6061. doi: 10.1371/journal.pone.0006061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Good L, Awasthi SK, Dryselius R, Larsson O, Nielsen PE. Bactericidal antisense effects of peptide-PNA conjugates. Nat Biotechnol. 2001;19:360–4. doi: 10.1038/86753. [DOI] [PubMed] [Google Scholar]
- 29.Gruegelsiepe H, Brandt O, Hartmann RK. Antisense inhibition of RNase P: mechanistic aspects and application to live bacteria. J Biol Chem. 2006;281:30613–20. doi: 10.1074/jbc.M603346200. [DOI] [PubMed] [Google Scholar]
- 30.Güell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–71. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 31.Han K, Kim KS, Bak G, Park H, Lee Y. Recognition and discrimination of target mRNAs by Sib RNAs, a cis-encoded sRNA family. Nucleic Acid Res. 2010 doi: 10.1093/nar/gkq292. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harth G, Horwitz MA, Tabatadze D, Zamecnik PC. Targeting the Mycobacterium tuberculosis 30/32-kDa mycolyl transferase complex as a therapeutic strategy against tuberculosis: Proof of principle by using antisense technology. Proc Natl Acad Sci USA. 2002;99:15614–9. doi: 10.1073/pnas.242612299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernández JA, Muro-Pastor AM, Flores E, Bes MT, Peleato ML, Fillat MF. Identification of a furA cis antisense RNA in the cyanobacterium Anabaena sp PCC 7120. J Mol Biol. 2006;355:325–34. doi: 10.1016/j.jmb.2005.10.079. [DOI] [PubMed] [Google Scholar]
- 34.Hernández JA, Pellicer S, Huang L, Peleato ML, Fillat MF. FurA modulates gene expression of alr3808, a DpsA homologue in Nostoc (Anabaena) sp PCC7120. FEBS Lett. 2007;581:1351–6. doi: 10.1016/j.febslet.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 35.Hjalt TA, Wagner EGH. Bulged-out nucleotides in an antisense RNA are required for rapid target RNA binding in vitro and inhibition in vivo. Nucleic Acid Res. 1995;23:580–7. doi: 10.1093/nar/23.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH. An antibody-based microarray assay for small RNA detection. Nucleic Acid Res. 2006;34:e52. doi: 10.1093/nar/gkl142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeon B, Zhang Q. Sensitization of Campylobacter jejuni to fluoroquinolone and macrolide antibiotics by antisense inhibition of the CmeABC multidrug efflux transporter. J Antimicrob Chemother. 2009;63:946–8. doi: 10.1093/jac/dkp067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji Y, Marra A, Rosenberg M, Woodnutt G. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J Bacteriol. 1999;181:6585–90. doi: 10.1128/jb.181.21.6585-6590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji Y, Yin D, Fox B, Holmes DJ, Payne D, Rosenberg M. Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol Lett. 2004;231:177–84. doi: 10.1016/S0378-1097(03)00931-5. [DOI] [PubMed] [Google Scholar]
- 40.Ji Y, Zhang B, Van Horn SF, Warren P, Woodnutt G, et al. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–9. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- 41.Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol Microbiol. 2007;64:738–54. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5′ and 3′ UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acid Res. 2005;33:1040–50. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawano M, Storz G, Rao BS, Rosner JL, Martin RG. Detection of low-level promoter activity within open reading frame sequences of Escherichia coli. Nucleic Acid Res. 2005;33:6268–76. doi: 10.1093/nar/gki928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kedar GC, Brown-Driver V, Reyes DR, Hilgers MT, Stidham MA, et al. Evaluation of the metS and murB loci for antibiotic discovery using targeted antisense RNA expression analysis in Bacillus anthracis. Antimicrob Agents Chemother. 2007;51:1708–18. doi: 10.1128/AAC.01180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JY, Cha HJ. Down-regulation of acetate pathway through antisense strategy in Escherichia coli: improved foreign protein production. Biotechnol Bioeng. 2003;83:841–53. doi: 10.1002/bit.10735. [DOI] [PubMed] [Google Scholar]
- 46.Kittle JD, Simons RW, Lee J, Kleckner N. Insertion sequence IS10 anti-sense pairing initiates by an interaction between the 5′ end of the target RNA and a loop in the anti-sense RNA. J Mol Biol. 1989;210:561–72. doi: 10.1016/0022-2836(89)90132-0. [DOI] [PubMed] [Google Scholar]
- 47.Kolb FA, Engdahl HM, Slagter-Jäger JG, Ehresmann B, Ehresmann C, et al. Progression of a loop-loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. EMBO J. 2000;19:5905–15. doi: 10.1093/emboj/19.21.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolb FA, Malmgren C, Westhof E, Ehresmann C, Ehresmann B, et al. An unusual structure formed by antisense-target RNA binding involves an extended kissing complex with a four-way junction and a side-by-side helical alignment. RNA. 2000;6:311–24. doi: 10.1017/s135583820099215x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolb FA, Westhof E, Ehresmann C, Ehresmann B, Wagner EGH, Romby P. Bulged residues promote the progression of a loop-loop interaction to a stable and inhibitory antisense-target RNA complex. Nucleic Acid Res. 2001;29:3145–53. doi: 10.1093/nar/29.15.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krinke L, Wulff DL. OOP RNA, produced from multicopy plasmids, inhibits lambda cII gene expression through an RNase III-dependent mechanism. Genes Dev. 1987;1:1005–13. doi: 10.1101/gad.1.9.1005. [DOI] [PubMed] [Google Scholar]
- 51.Krinke L, Wulff DL. RNase III-dependent hydrolysis of l cII-O gene mRNA mediated by l OOP antisense RNA. Genes Dev. 1990;4:2223–33. doi: 10.1101/gad.4.12a.2223. [DOI] [PubMed] [Google Scholar]
- 52.Landt SG, Abeliuk E, McGrath PT, Lesley JA, McAdams HH, Shapiro L. Small non-coding RNAs in Caulobacter crescentus. Mol Microbiol. 2008;68:600–14. doi: 10.1111/j.1365-2958.2008.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee EJ, Groisman EA. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol. 2010;76:1020–33. doi: 10.1111/j.1365-2958.2010.07161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao SM, Wu TH, Chiang CH, Susskind MM, McClure WR. Control of gene expression in bacteriophage P22 by a small antisense RNA. I. Characterization in vitro of the Psar promoter and the sar RNA transcript. Genes Dev. 1987;1:197–203. doi: 10.1101/gad.1.2.197. [DOI] [PubMed] [Google Scholar]
- 55.Liu JM, Livny J, Lawrence MS, Kimball MD, Waldor MK, Camilli A. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acid Res. 2009;37:e46. doi: 10.1093/nar/gkp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livny J, Waldor MK. Identification of small RNAs in diverse bacterial species. Curr Opin Microbiol. 2007;10:96–101. doi: 10.1016/j.mib.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 57.López-Gomollón S, Hernández JA, Wolk CP, Peleato ML, Fillat MF. Expression of furA is modulated by NtcA and strongly enhanced in heterocysts of Anabaena sp PCC 7120. Microbiology. 2007;153:42–50. doi: 10.1099/mic.0.2006/000091-0. [DOI] [PubMed] [Google Scholar]
- 58.Malmgren C, Wagner EGH, Ehresmann C, Ehresmann B, Romby P. Antisense RNA control of plasmid R1 replication. The dominant product of the antisense rna-mrna binding is not a full RNA duplex. J Biol Chem. 1997;272:12508–12. doi: 10.1074/jbc.272.19.12508. [DOI] [PubMed] [Google Scholar]
- 59.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35:962–74. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Massé E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10:140–5. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 62.Mellbye BL, Weller DD, Hassinger JN, Reeves MD, Lovejoy CE, et al. Cationic phosphorodiamidate morpholino oligomers efficiently prevent growth of Escherichia coli in vitro and in vivo. J Antimicrob Chemother. 2010;65:98–106. doi: 10.1093/jac/dkp392. [DOI] [PubMed] [Google Scholar]
- 63.Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, et al. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One. 2009;4:e7526. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitev GM, Mellbye BL, Iversen PL, Geller BL. Inhibition of intracellular growth of Salmonella enterica serovar Typhimurium in tissue culture by antisense peptide-phosphorodiamidate morpholino oligomer. Antimicrob Agents Chemother. 2009;53:3700–4. doi: 10.1128/AAC.00099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morrissey JA, Cockayne A, Hill PJ, Williams P. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect Immun. 2000;68:6281–8. doi: 10.1128/iai.68.11.6281-6288.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakashima N, Tamura T, Good L. Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli. Nucleic Acid Res. 2006;34:e138. doi: 10.1093/nar/gkl697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nekhotiaeva N, Awasthi SK, Nielsen PE, Good L. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol Ther. 2004;10:652–9. doi: 10.1016/j.ymthe.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 68.Nikravesh A, Dryselius R, Faridani OR, Goh S, Sadeghizadeh M, et al. Antisense PNA accumulates in Escherichia coli and mediates a long post-antibiotic effect. Mol Ther. 2007;15:1537–42. doi: 10.1038/sj.mt.6300209. [DOI] [PubMed] [Google Scholar]
- 69.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186:6698–705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, et al. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36:1913–27. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res Microbiol. 2009;160:278–87. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 72.Papoutsakis ET. Engineering solventogenic clostridia. Curr Opin Biotechnol. 2008;19:420–9. doi: 10.1016/j.copbio.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 73.Parish CA, de la Cruz M, Smith SK, Zink D, Baxter J, et al. Antisense-guided isolation and structure elucidation of pannomycin, a substituted cis-decalin from Geomyces pannorum. J Nat Prod. 2009;72:59–62. doi: 10.1021/np800528a. [DOI] [PubMed] [Google Scholar]
- 74.Passalacqua KD, Varadarajan A, Ondov BD, Okou DT, Zwick ME, Bergman NH. Structure and complexity of a bacterial transcriptome. J Bacteriol. 2009;191:3203–11. doi: 10.1128/JB.00122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perocchi F, Xu Z, Clauder-Münster S, Steinmetz LM. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acid Res. 2007;35:e128. doi: 10.1093/nar/gkm683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perret S, Maamar H, Bélaich JP, Tardif C. Use of antisense RNA to modify the composition of cellulosomes produced by Clostridium cellulolyticum. Mol Microbiol. 2004;51:599–607. doi: 10.1046/j.1365-2958.2003.03860.x. [DOI] [PubMed] [Google Scholar]
- 77.Persson C, Wagner EGH, Nordström K. Control of replication of plasmid R1: kinetics of in vitro interaction between the antisense RNA, CopA, and its target, CopT. EMBO J. 1988;7:3279–88. doi: 10.1002/j.1460-2075.1988.tb03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Persson C, Wagner EGH, Nordström K. Control of replication of plasmid R1: formation of an initial transient complex is rate-limiting for antisense RNA--target RNA pairing. EMBO J. 1990;9:3777–85. doi: 10.1002/j.1460-2075.1990.tb07591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Persson C, Wagner EGH, Nordström K. Control of replication of plasmid R1: structures and sequences of the antisense RNA, CopA, required for its binding to the target RNA, CopT. EMBO J. 1990;9:3767–75. doi: 10.1002/j.1460-2075.1990.tb07590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci USA. 2009;106:15406–11. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Predki PF, Nayak LM, Gottlieb MB, Regan L. Dissecting RNA-protein interactions: RNA-RNA recognition by Rop. Cell. 1995;80:41–50. doi: 10.1016/0092-8674(95)90449-2. [DOI] [PubMed] [Google Scholar]
- 82.Rasmussen LC, Sperling-Petersen HU, Mortensen KK. Hitting bacteria at the heart of the central dogma: sequence-specific inhibition. Microb Cell Fact. 2007;6:24. doi: 10.1186/1475-2859-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rasmussen S, Nielsen HB, Jarmer H. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol. 2009;73:1043–57. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Recalcati S, Minotti G, Cairo G. Iron regulatory proteins: from molecular mechanisms to drug development. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2009.2983. in press. [DOI] [PubMed] [Google Scholar]
- 85.Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr Biol. 2001;11:1369–73. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 86.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acid Res. 2004;32:3340–53. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schlüter JP, Reinkensmeier J, Daschkey S, Evguenieva-Hackenberg E, Janssen S, et al. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti. BMC Genomics. 2010;11:245. doi: 10.1186/1471-2164-11-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Selinger DW, Cheung KJ, Mei R, Johansson EM, Richmond CS, et al. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol. 2000;18:1262–8. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 89.Sharma CM, Hoffmann S, Darfeuille F, Findeiß S, Sittka A, et al. The primary transcriptome of the major human pathogen, Helicobacter pylori. Nature. 2010;464:250–5. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 90.Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–45. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Silby MW, Levy SB. Use of in vivo expression technology to identify genes important in growth and survival of Pseudomonas fluorescens Pf0-1 in soil: discovery of expressed sequences with novel genetic organization. J Bacteriol. 2004;186:7411–9. doi: 10.1128/JB.186.21.7411-7419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Silby MW, Levy SB. Overlapping protein-encoding genes in Pseudomonas fluorescens Pf0-1. PLoS Genet. 2008;4:e1000094. doi: 10.1371/journal.pgen.1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sillers R, Al-Hinai MA, Papoutsakis ET. Aldehyde-alcohol dehydrogenase and/or thiolase overexpression coupled with CoA transferase downregulation lead to higher alcohol titers and selectivity in Clostridium acetobutylicum fermentations. Biotechnol Bioeng. 2009;102:38–49. doi: 10.1002/bit.22058. [DOI] [PubMed] [Google Scholar]
- 94.Silvaggi JM, Perkins JB, Losick R. Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis. J Bacteriol. 2006;188:532–41. doi: 10.1128/JB.188.2.532-541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Simons RW, Kleckner N. Translational control of IS10 transposition. Cell. 1983;34:683–91. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- 96.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Steglich C, Futschik ME, Lindell D, Voss B, Chisholm SW, Hess WR. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e1000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stork M, Di Lorenzo M, Welch TJ, Crosa JH. Transcription termination within the iron transport-biosynthesis operon of Vibrio anguillarum requires an antisense RNA. J Bacteriol. 2007;189:3479–88. doi: 10.1128/JB.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stougaard P, Molin S, Nordström K. RNAs involved in copy-number control and incompatibility of plasmid R1. Proc Natl Acad Sci USA. 1981;78:6008–12. doi: 10.1073/pnas.78.10.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thisted T, Sørensen NS, Wagner EGH, Gerdes K. Mechanism of post-segregational killing: Sok antisense RNA interacts with Hok mRNA via its 5′-end single-stranded leader and competes with the 3′-end of Hok mRNA for binding to the mok translational initiation region. EMBO J. 1994;13:1960–8. doi: 10.1002/j.1460-2075.1994.tb06465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tilley LD, Mellbye BL, Puckett SE, Iversen PL, Geller BL. Antisense peptide-phosphorodiamidate morpholino oligomer conjugate: dose-response in mice infected with Escherichia coli. J Antimicrob Chemother. 2007;59:66–73. doi: 10.1093/jac/dkl444. [DOI] [PubMed] [Google Scholar]
- 102.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–6. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 103.Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984;38:861–70. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- 104.Tomizawa J. Control of ColE1 plasmid replication. Interaction of Rom protein with an unstable complex formed by RNA I and RNA II. J Mol Biol. 1990;212:695–708. doi: 10.1016/0022-2836(90)90231-a. [DOI] [PubMed] [Google Scholar]
- 105.Tomizawa J, Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci USA. 1981;78:6096–100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tomizawa J, Itoh T, Selzer G, Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci USA. 1981;78:1421–5. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tomizawa J, Som T. Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell. 1984;38:871–8. doi: 10.1016/0092-8674(84)90282-4. [DOI] [PubMed] [Google Scholar]
- 108.Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol Microbiol. 2008;70:965–82. doi: 10.1111/j.1365-2958.2008.06458.x. [DOI] [PubMed] [Google Scholar]
- 109.Tummala SB, Junne SG, Papoutsakis ET. Antisense RNA downregulation of coenzyme A transferase combined with alcohol-aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J Bacteriol. 2003;185:3644–53. doi: 10.1128/JB.185.12.3644-3653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tummala SB, Junne SG, Paredes CJ, Papoutsakis ET. Transcriptional analysis of product-concentration driven changes in cellular programs of recombinant Clostridium acetobutylicumstrains. Biotechnol Bioeng. 2003;84:842–54. doi: 10.1002/bit.10851. [DOI] [PubMed] [Google Scholar]
- 111.Tummala SB, Welker NE, Papoutsakis ET. Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J Bacteriol. 2003;185:1923–34. doi: 10.1128/JB.185.6.1923-1934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jäger JG, et al. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acid Res. 2003;31:6435–43. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wagner EGH, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv Genet. 2002;46:361–98. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- 114.Wagner EGH, Blomberg P, Nordström K. Replication control in plasmid R1: duplex formation between the antisense RNA, CopA, and its target, CopT, is not required for inhibition of RepA synthesis. EMBO J. 1992;11:1195–203. doi: 10.1002/j.1460-2075.1992.tb05160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wagner EGH, Simons RW. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol. 1994;48:713–42. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 116.Wang Q, Harshey RM. Rcs signalling-activated transcription of rcsA induces strong anti-sense transcription of upstream fliPQR flagellar genes from a weak intergenic promoter: regulatory roles for the anti-sense transcript in virulence and motility. Mol Microbiol. 2009;74:71–84. doi: 10.1111/j.1365-2958.2009.06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–51. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–28. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu TH, Liao SM, McClure WR, Susskind MM. Control of gene expression in bacteriophage P22 by a small antisense RNA. II. Characterization of mutants defective in repression. Genes Dev. 1987;1:204–12. doi: 10.1101/gad.1.2.204. [DOI] [PubMed] [Google Scholar]
- 120.Xiao B, Li W, Guo G, Li B, Liu Z, et al. Identification of small noncoding RNAs in Helicobacter pylori by a bioinformatics-based approach. Curr Microbiol. 2009;58:258–63. doi: 10.1007/s00284-008-9318-2. [DOI] [PubMed] [Google Scholar]
- 121.Yachie N, Numata K, Saito R, Kanai A, Tomita M. Prediction of non-coding and antisense RNA genes in Escherichia coli with Gapped Markov Model. Gene. 2006;372:171–81. doi: 10.1016/j.gene.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 122.Zemanová M, Kaderábková P, Pátek M, Knoppová M, Silar R, Nesvera J. Chromosomally encoded small antisense RNA in Corynebacterium glutamicum. FEMS Microbiol Lett. 2008;279:195–210. doi: 10.1111/j.1574-6968.2007.01024.x. [DOI] [PubMed] [Google Scholar]
- 123.Zhang C, Ondeyka JG, Zink DL, Basilio A, Vicente F, et al. Isolation, structure and antibacterial activity of pleosporone from a pleosporalean ascomycete discovered by using antisense strategy. Bioorg Med Chem. 2009;17:2162–6. doi: 10.1016/j.bmc.2008.04.018. [DOI] [PubMed] [Google Scholar]