Chromatin signatures of the Drosophila replication program (original) (raw)
Abstract
DNA replication initiates from thousands of start sites throughout the Drosophila genome and must be coordinated with other ongoing nuclear processes such as transcription to ensure genetic and epigenetic inheritance. Considerable progress has been made toward understanding how chromatin modifications regulate the transcription program; in contrast, we know relatively little about the role of the chromatin landscape in defining how start sites of DNA replication are selected and regulated. Here, we describe the Drosophila replication program in the context of the chromatin and transcription landscape for multiple cell lines using data generated by the modENCODE consortium. We find that while the cell lines exhibit similar replication programs, there are numerous cell line-specific differences that correlate with changes in the chromatin architecture. We identify chromatin features that are associated with replication timing, early origin usage, and ORC binding. Primary sequence, activating chromatin marks, and DNA-binding proteins (including chromatin remodelers) contribute in an additive manner to specify ORC-binding sites. We also generate accurate and predictive models from the chromatin data to describe origin usage and strength between cell lines. Multiple activating chromatin modifications contribute to the function and relative strength of replication origins, suggesting that the chromatin environment does not regulate origins of replication as a simple binary switch, but rather acts as a tunable rheostat to regulate replication initiation events.
With every cell division, DNA replication must initiate in a coordinated manner from thousands of start sites or origins of DNA replication. Potential origins of DNA replication are selected by the heterohexameric origin recognition complex (ORC), which is conserved in all eukaryotes. The replicative helicase, MCM2-7 complex, is loaded at ORC-binding sites in G1 to form the pre-replicative complex (pre-RC) (for review, see Bell and Dutta 2002). In S. cerevisiae, ORC recognizes a degenerate consensus sequence, the ACS (ARS consensus sequence) (Marahrens and Stillman 1992), which is necessary, but not sufficient for ORC binding (Breier et al. 2004). In higher eukaryotes, a defined consensus sequence has yet to emerge, suggesting that additional features such as chromatin organization and modification will be critical components for defining sequences that will function as origins of replication.
The assembly, organization, and modification of histone octamers on the DNA regulate the accessibility of the DNA to _trans_-acting factors such as transcription factors and RNA polymerase II (RNA Pol II) (for review, see Rando and Chang 2009). Promoter elements have a specific nucleosome organization with a nucleosome-free region immediately upstream of the transcription start site (TSS) and well-positioned nucleosomes within the gene body (Lee et al. 2007). Similarly, a number of epigenetic histone modifications modulate the recruitment of transcription factors to DNA and gene-expression levels in what is often referred to as the histone code hypothesis (Jenuwein and Allis 2001). While much progress has been made in our understanding of how chromatin structure and organization regulate gene expression, we know comparatively little about their contribution to the DNA replication program.
In S. cerevisiae, the organization of nucleosomes is an important determinant of ORC localization. ORC localizes to nucleosome-free regions and is required to precisely position nucleosomes flanking the origin of replication (Berbenetz et al. 2010; Eaton et al. 2010). This precise nucleosome organization is required for origin function as occluding the ACS within a nucleosome or moving the upstream flanking nucleosome abrogates origin function (Simpson 1990; Lipford and Bell 2001). In Drosophila, nucleosome organization also appears to be a defining feature of ORC-binding sites. Sites of ORC enrichment are depleted for bulk nucleosomes and enriched for the histone variant H3.3 (MacAlpine et al. 2010). Recent experiments profiling Drosophila nucleosome turnover in near real time by “covalent attachment of tags to capture histones and identify turnover” (CATCH-IT) found that ORC-associated sites undergo active nucleosome exchange (Deal et al. 2010). Together, these data suggest that in both yeast and higher eukaryotes, a primary determinant of ORC localization is the accessibility of the DNA in higher order chromatin.
Histone acetylation has been positively correlated with replication origin activity in a variety of experimental systems. In S. cerevisiae, deletion of the histone deacetylase, RPD3, results in the earlier activation of a subset of late-firing origins of replication (Vogelauer et al. 2002; Knott et al. 2009). Similar experiments in Drosophila have shown that replication initiation during the developmentally programmed amplification of the chorion locus is also sensitive to changes in local histone acetylation (Aggarwal and Calvi 2004). In mammalian cells, differences in local chromatin acetylation at the beta-globin locus between erythroid and nonerythroid cells are reflected in the activity of the beta-globin replication origin (Goren et al. 2008). Finally, the loading of a transcriptional activator, GAL4-VP16, on a plasmid is sufficient to localize origin activity in Xenopus extracts (Danis et al. 2004). This effect is not dependent on active transcription, but instead is correlated with the local acetylation of histones at the site of initiation. The role of chromatin modification in the selection of origins of replication is less clear, although recent studies at a select number of mammalian origins of replication indicate that HBO1 (also known as MYST2), a histone H4 acetylase, interacts with CDT1 and is required for the subsequent loading of the MCM complex to form the pre-RC (Miotto and Struhl 2010).
To better understand how the DNA replication program is established and regulated, we have examined multiple DNA replication data sets in the context of the diverse data types generated by the modENCODE (model organism encyclopedia of DNA elements) consortium (Celniker et al. 2009). The goal of the modENCODE project is to identify all functional DNA elements in the genomes of the model organisms D. melanogaster and C. elegans. Together, the consortium has generated nearly 1000 genomic data sets, consisting of array and sequencing-based experiments that describe the transcription program, map the chromatin landscape, identify transcription factor and insulator binding sites, and characterize the DNA replication program across multiple cell lines and developmental stages (The modENCODE Consortium 2010).
Our analysis of the similarities and differences in the DNA replication program from three Drosophila cell lines has provided unique insights into the role of chromatin organization and modifications in regulating the DNA replication program. Genome-wide replication timing and early origins of replication are correlated with a variety of activating chromatin marks. Similarly, we find that ORC-binding sites are enriched for a subset of these chromatin marks as well as additional DNA-binding proteins, suggesting that the selection and regulation of origins may be controlled by distinct factors. We were unable to identify a simple consensus sequence analogous to the yeast ACS from the high-resolution ChIP-seq ORC-binding data. Instead, we were able to use machine-learning approaches to classify ORC-binding sites based on sequence features, chromatin modifications, and DNA-binding proteins. Finally, we generated accurate computational models to predict origin function based on the chromatin landscape.
Results
Characterization of the Drosophila replication program in three cell lines
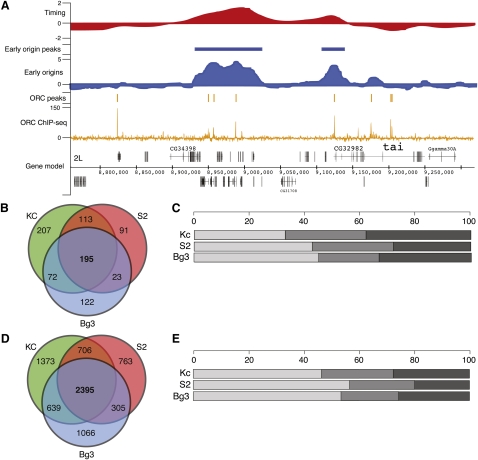
As part of the modENCODE consortium, we have utilized multiple genomic approaches to characterize the Drosophila DNA replication program in three different cell lines. Two of these cell lines are of embryonic origin (Kc167 [Kc] and S2-DRSC [S2]), and the third is derived from neuronal tissue (ML-DmBG3-c2 [Bg3]); together, they represent some of the most commonly utilized cell lines in the Drosophila research community. We have taken a top-down approach, characterizing the replication program by utilizing increasingly high-resolution and complementary assays (Fig. 1A). Specifically, we used genomic tiling arrays to determine the relative time of replication for all unique sequences in the Drosophila genome (Fig. 1A, top), map early activating origins of replication (Fig. 1A, middle), and identify, at near nucleotide resolution, the locations of ORC binding by using high-throughput sequencing (Fig. 1A, bottom).
Figure 1.
The Drosophila replication program across three cell lines. (A) Replication program in S2-DRSC cells. Genome browser track of whole-genome S-phase replication timing profiles as the log2 ratio of early to late replicating sequences (red), early origin activity as the log2 ratio of BrdU enrichment to input DNA (blue), ORC-binding sites as input corrected ChIP-seq tag depth (orange), and gene models for a 500-kb region of chromosome 2L. (B) Overlap of early origins in three cell lines. The Venn diagram shows the overlap in total early origin peaks from each cell line. (C) Distribution of early origin meta-peaks per cell line. The percentage of early origin peaks found in three cell lines (light gray), two cell lines (medium gray), or one cell line (dark gray). (D) Overlap of ORC ChIP-seq peaks in three cell lines. As in B, the Venn diagram depicts the overlap in ORC peaks for each cell line. (E) Distribution of ORC meta-peaks per cell line. Same as C for ORC ChIP-seq peaks.
Genomic tiling arrays were used to identify the relative time of DNA replication during S phase for unique sequences in the Drosophila genome. Briefly, synchronized cells were pulse labeled with the nucleotide analog, 5-bromo-2-deoxyuridine (BrdU), during either early or late S phase, resulting in the differential labeling of early and late replicating sequences (MacAlpine et al. 2004). These fractions of early and late replicating sequences were then hybridized to genomic tiling arrays to determine the relative time of replication. We found that the relative time of DNA replication was correlated across the three cell lines (Table 1). The replication timing of the X chromosome is one notable exception; it completes replication significantly earlier than the autosomes in the male cell lines (Schwaiger et al. 2009; L DeNapoli, M Eaton, and D MacAlpine, in prep.).
Table 1.
Pearson correlation of whole-genome replication timing between cell lines
Early origins of replication were identified in the three cell lines by pulse labeling cells with BrdU in the presence of hydroxyurea (HU), a potent inhibitor of nucleotide biosynthesis. HU treatment results in stalled replication forks and the activation of the intra-S-phase checkpoint (Santocanale and Diffley 1998; Shirahige et al. 1998). Thus, only those sequences immediately adjacent to early activating origins of replication will incorporate BrdU, allowing the enrichment of early origin proximal sequences by immunoprecipitation with anti-BrdU antibodies (MacAlpine et al. 2004). We identified 630, 457, and 433 early origins of replication in Kc, S2, and Bg3 cells, respectively. To facilitate inter-cell line comparisons, we assembled a single set of 823 genomic loci that had evidence for early origin activity in at least one of the three cell lines. We will refer to this as the set of early origin meta-peaks, and it represents locations in the Drosophila genome that are capable of functioning as a replication origin. Almost a quarter of these early origin meta-peaks (195) were found in all three cell lines and approximately half (403) were found in at least two cell lines (Fig. 1B). For each of the three cell lines, at least two-thirds of the early origins were also found in another cell line (Fig. 1C). Importantly, 82% of the early origin meta-peaks contained an ORC-associated sequence (P ≤ 1 × 10−5, bootstrap R = 100,000; see Methods).
Recent advances in sequencing technology have allowed the use of high-throughput sequencing to directly sequence DNA from chromatin immunoprecipitation experiments (ChIP-seq) (Johnson et al. 2007; Robertson et al. 2007). We have used ChIP-seq to analyze the genome-wide distribution of ORC. ChIP-seq of biological replicates from independent experiments was performed on each of the cell lines. We identified 5159, 4230, and 4477 distinct ORC-binding sites in Kc, S2, and Bg3 cells, respectively. We then generated a set of ORC meta-peaks analogous to the set of early origin meta-peaks described above. We identified 7246 ORC meta-peaks, of which more than a third (2395) have support in all three cell lines, and more than half of these sites (4045) were found in at least two cell lines (Fig. 1D). Examination of each of the individual cell lines revealed almost 75% of the ORC peaks identified in a particular cell line were also found in at least one other cell line (Fig. 1E).
Diverse chromatin marks define the replication program
The chromatin landscape clearly impacts both the expression and the replication of the genome. For example, the transcriptionally active euchromatin typically replicates prior to the repressed heterochromatic sequences (for review, see Gilbert 2010). Studies in yeast, Drosophila, and mammalian systems have shown that changes in histone acetylation (Aggarwal and Calvi 2004; Goren et al. 2008; Knott et al. 2009) are associated with changes in the replication program. However, a comprehensive view of the replication program in the context of chromatin modifications and DNA-binding proteins is lacking.
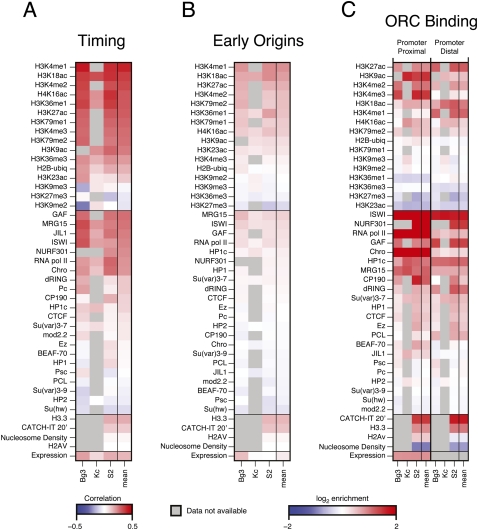
The different modENCODE data types across multiple cell lines (The modENCODE Consortium 2010) allowed us to define the chromatin and transcription landscape associated with features of the DNA replication program. For each replication data type (replication timing, early origins, and ORC binding), we generated a 43 × 3 matrix, with each column representing a specific cell line and each row representing the enrichment or correlations with chromatin marks, DNA-binding proteins, nucleosome density, histone variants, nucleosome turnover (CATCH-IT) (Deal et al. 2010), and gene expression (RNA-seq) (Fig. 2). For the replication timing profiles where we did not have discrete peak calls, we calculated the Spearman's correlation between each factor with the whole-genome replication timing profile (Fig. 2A). For early origins of replication (Fig. 2B) and ORC-binding sites (Fig. 2C), we calculated the median log2 enrichment of each factor within all BrdU peaks and within 500 bp of ORC ChIP-seq peak centers, respectively. The rightmost column of each matrix is a summary column depicting the average signal or correlation for each factor derived from the three independent cell lines.
Figure 2.
The chromatin landscape of the replication program. (A) Chromatin correlations with replication timing. The genome-wide replication timing profile of each cell line was paired with the genome-wide array scores for each chromatin factor, and the pairwise correlation of the factor with replication timing was computed (Spearman's ρ). The correlation ρ ranges from −0.5 (blue) to +.5 (red). (B) The chromatin landscape of early origins. The log2 enrichment for each factor within early origin peaks was determined for each cell line. The enrichment ranges from −2 (blue) to +2 (red). (C) The chromatin landscape of ORC-binding sites. ORC-associated sequences were divided into TSS proximal (overlapping a TSS) and TSS distal (not overlapping a TSS). The log2 enrichment for each factor within 500 bp of the ORC peak centers was determined for each cell line. The enrichment ranges from −2 (blue) to +2 (red). In all panels, gray boxes represent an experiment that has not yet been submitted to modENCODE. See Methods for details.
We found that the selection and regulation of DNA replication origins is associated with distinct sets of chromatin marks and DNA-binding proteins. Prior studies have associated early replication with active transcription and the presence of “activating” chromatin modifications such as histone acetylation, whereas late replication is associated with “repressive” chromatin marks such as those found in the heterochromatin (for review, see Gilbert 2010). Indeed, we found that gene expression is positively correlated with replication timing, as are generally euchromatic marks such as H3K4me1 and H3K18ac (Fig. 2A). In contrast, heterochromatic marks such as H3K27me3 and H3K9me2 are negatively correlated with replication timing. The sequences surrounding early origins were also enriched for activating chromatin marks as well as specific DNA-binding proteins, including chromatin remodeling factors (Fig. 2B).
Because many of the ORC-binding sites colocalized with promoters of active genes (MacAlpine et al. 2010), we separated the ORC-binding sites into those that are TSS proximal (within 1 kb of a TSS) and those that were not at a TSS (distal). We were particularly interested in chromatin features that are shared between ORC-binding sites both proximal and distal to promoters. Additionally, marks that are specific to ORC sites distal from a promoter will be of interest, as these marks may be required for ORC binding or function in the absence of a promoter.
ORC-binding sites proximal to TSSs were enriched for chromatin remodelers such as the NURF complex (NURF301 [also known as E(BX)], ISWI) as well as other DNA-binding proteins such as GAF, RNA Pol II, and CHRO (Fig. 2C). These TSS-associated ORC sites were also enriched for H3K9ac, H3K27ac, H3K4me2, and H3K4me3—marks frequently found at promoters. Interestingly, those ORC sites that did not overlap with a TSS (distal) were also enriched for chromatin remodelers ISWI and NURF301, as well as GAF, which has also been implicated in chromatin remodeling (Petesch and Lis 2008). Consistent with the idea of ORC localizing to dynamic and active chromatin, we found an enrichment for CATCH-IT and H3.3 at ORC sites both proximal and distal to TSSs, as well as a reduction in bulk nucleosome occupancy (The modENCODE Consortium 2010). ORC sites not located at promoters were enriched for many of the same histone marks as those at promoters, with a few notable exceptions. We found a decrease in H3K4me3 at ORC sites distal from a promoter, as well as an increase in H3K18ac and H3K4me1.
Chromatin features specific to transcription start sites such as RNA Pol II and H2Av were decreased at ORC-binding sites distal to promoter elements. A small amount of RNA Pol II signal remained in the TSS distal ORC-binding sites; however, in comparison to the local enrichment of ISWI and GAF, there was a clear reduction in local signal (Supplemental Fig. S1). The remaining signal may be due to unannotated transcription start sites.
Chromatin marks that are associated with active transcription through gene bodies (e.g., H3K79me1, H3K36me1, and H3K36me3) were not found above background levels at any ORC-binding sites. However, H3K36me1 was found specifically flanking those ORC-binding sites that did not coincide with a TSS (The modENCODE Consortium 2010). ORC has been shown to facilitate the formation of heterochromatin and HP1 binding (Pak et al. 1997); however, we found that ORC sites were depleted for heterochromatic histone modifications such as H3K27me3 and H3K9me2/3 and were only slightly enriched for HP1. This may be due, in part, to the inability to map distinct ORC-binding sites in repetitive sequences, a current limitation of high-throughput sequencing approaches.
We also examined the chromatin signatures of promoter elements with and without ORC associated to determine whether there were unique chromatin signatures specific for ORC associated promoters. Since those promoters with proximal ORC binding tend to be far more actively transcribed than those without ORC (Supplemental Fig. S2), we limited our comparison to active promoter elements only. We found that ORC-associated promoters had modestly increased chromatin remodeling activities, decreased nucleosome occupancy, and greater evidence of nucleosome turn-over relative to other active promoters not associated with ORC (Supplemental Fig. S3). In summary, these results indicate that dynamic chromatin environments may contribute to ORC localization and the subsequent activation of replication origins.
Potential _cis_- and _trans_-acting elements directing ORC association
ORC purified from higher eukaryotes exhibits little if any sequence specificity in vitro (Vashee et al. 2003; Remus et al. 2004). Despite the apparent lack of specificity, we found that ORC localized to specific chromosomal locations in three different cell lines. In our prior ORC ChIP-chip experiments (MacAlpine et al. 2010), we were unable to identify a single sequence motif that was predictive for ORC binding, but rather, we were able to identify sequence words (_k_-mers) that in combination were able to accurately classify sequences as ORC associated or not using support vector machines (SVMs). We revisited the issue of specific sequences directing ORC localization with our ORC-binding data derived from the higher resolution ChIP-seq data sets.
First, we sought to identify potential _cis_-acting motif elements in those sequences that were associated with ORC. We identified 3416 sequences representing the intersection of peaks in each of the three cell lines. The intersection of the ORC peaks was used rather than the previously described meta-peaks to increase the nucleotide level resolution. The center of the intersection was used as the epicenter of the ORC-binding sites and extended 250 bp up- and downstream to yield a 500-bp fragment. Using the motif identification tool MEME (Bailey and Elkan 1994), we were only able to identify simple repetitive elements (CA)n, (CT)n, and (CG)n that, although enriched (Supplemental Fig. S4), were not particularly predictive of ORC binding (Supplemental Fig. S5).
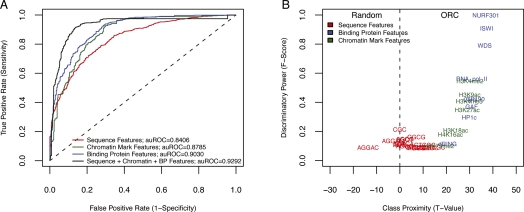
We decided to build upon our earlier success using support vector machines (SVM) to classify ORC-binding sites based on primary sequence (MacAlpine et al. 2010) by including additional information such as chromatin modifications and DNA-binding proteins. An SVM is a classification algorithm that is trained on a set of labeled samples represented as feature vectors (e.g., ORC bound or unbound sequences), and after training is able to accurately distinguish between samples in a test data set. In accordance with this, we generated a feature vector describing each ORC-binding site and an equal number of random loci using unique sequence _k_-mers (where k is 1–6, excluding reverse complements), 15 chromatin marks, and 20 DNA-binding proteins, for a total of 2765 features from the S2 data set. Given the input training set, which consisted of an equal number of ORC bound and random loci (balanced for promoter occupancy) from chromosomes 2L, 3L, and 3R, we generated an SVM model using 10-fold cross-validation of the training data set. The X and fourth chromosomes possess unique chromatin environments and were excluded from the training and test data sets. Specifically, the single male X chromosome is hyperacetylated on H4K16 (Lucchesi et al. 2005) and the 1.2-Mb fourth chromosome has a high-transposon density and a prevalence of repressive heterochromatin marks (Riddle et al. 2009).
After training the SVM, we identified those features with the most discriminatory power (_F_-score > 0.075) and thus reduced the length of our feature vector from 2765 features to only 34 features. The model generated from this reduced feature set was then used to classify ORC-bound sequences on chromosome 2R, which was deliberately left out of the training. The results of the testing are presented as receiver operator characteristic (ROC) curves, with the _y_-axis representing the sensitivity of the assay (true-positive rate) as a function of 1-specificity (false-positive rate). We found that we could accurately predict ORC-binding sites using either sequence, chromatin modifications, or DNA-binding proteins (Fig. 3A). However, combining the different features resulted in a marked increase in accuracy of the predictions. For example, at a sensitivity of 83% the false-positive rate was ∼8%. Although there were fewer chromatin factors available for the other two cell lines, the SVM was still able to accurately classify origins in both Kc and Bg3 (Supplemental Fig. S6).
Figure 3.
Sequence, chromatin, and DNA-binding proteins classify ORC-binding sites. (A) SVM performance was gauged by the ROC curve resulting from separately using sequence features, chromatin mark features, binding protein features, or a combination of all three. In each case, the SVM was trained using 10-fold cross validation on three chromosome arms (2L, 3L, and 3R) and tested on a fourth chromosome arm (2R). (B) The importance of individual features was determined by plotting the _F_-score as a function of class proximity represented here as a _t_-statistic.
To better understand the specific features that contributed to the discriminatory accuracy of the SVM, we plotted the class proximity (defined as a _t_-statistic obtained from comparing the feature counts between positive and negative sets) for each feature against the _F_-score (Fig. 3B). In each case, the _t_-statistic denoted the class to which each feature most correlated, and the discriminatory power indicated its overall importance in classifying ORC sites. Our analysis showed that binding proteins were by far the most discriminative feature type, followed closely by chromatin marks. Sequence features were neither very discriminative nor strongly correlated to either class. This analysis was therefore able to identify those features most highly correlated with ORC-binding sites across all cell lines (Supplemental Fig. S10), namely, WDS (which interacts with complexes whose functions include histone acetylation and enabling ISWI to remodel chromatin) (Suganuma et al. 2008), chromatin remodelers (NURF301, ISWI) and “activating” chromatin modifications (H3K4me2, H3K4me3, H3K27ac, etc.).
Predicting early origin usage from the chromatin landscape
Genome-wide and locus-specific experiments have revealed that early replicating regions of the genome typically correlate with “activating” chromatin marks and that changing the local acetylation pattern of surrounding sequences can modulate replication activity (Aggarwal and Calvi 2004; The ENCODE Project Consortium 2007; Karnani et al. 2007; Knott et al. 2009). However, genome-wide experiments have simply revealed global correlations and the observations taken from locus-specific examples may not apply to all origins on a genome-wide scale. Finally, it is difficult to discern specific and nonspecific effects from altering global patterns of histone acetylation. For example, deletion of the global histone deacetylase RPD3 in yeast results in the earlier activation of a subset of late-firing origins (Knott et al. 2009); however, the global reduction in histone H3 acetylation levels may also impact the transcriptional control of key replication initiation factors.
The chromatin modENCODE data sets derived from multiple cell lines (Kharchenko et al. 2011) provided a unique opportunity to identify chromatin factors and DNA-binding proteins that might distinguish differential origin usage or ORC occupancy between cell lines. Using S2 and Bg3 cells, we first identified ORC-binding sites that were only found in Bg3 cells, but not S2 cells. Comparison of the chromatin signatures between Bg3 and S2 cells for those locations that were only bound by ORC in Bg3 cells revealed only subtle changes in chromatin factors between the cell lines (Supplemental Fig. S7A). For example, very modest decreases were observed for ISWI and RNA Pol II in S2 cells at the ORC locations that were only utilized in Bg3 cells. Similar modest changes were also observed between S2 and Bg3 cells when ORC locations specific to S2 cells were examined.
We also examined the chromatin signatures of early origins of replication in a similar manner (Supplemental Fig. S7B). Again, early origins specific to Bg3 cells exhibited only a subtle difference in chromatin signatures between Bg3 and S2 cells. A slight increase in repressive heterochromatin marks (H3K9me2 and H3K9me3) was observed at those sites in S2 cells. More dramatic differences in chromatin factors were observed for early origins specific to S2 cells. Specifically, a loss of chromatin remodelers, RNA Pol II, and activating marks were observed in Bg3 cells at those locations. Together, these results suggest that early origins specific to S2 cells are likely defined by increased activating chromatin marks, RNA Pol II, and chromatin remodeling activities. However, Bg3-specific origins of replication exhibited much more subtle differences in the local chromatin environment between cell lines, suggesting that even minor changes in chromatin environment may impact origin usage.
The fact that there was not a single chromatin mark or DNA-binding protein that was highly correlative with origin function by itself prompted us to test the hypothesis that perhaps a complex integration of diverse chromatin marks and DNA-binding proteins was defining a local chromatin “terroir” favorable to origin usage. Our goal was to develop a predictive model of origin function based on chromatin and DNA-binding protein input. Specifically, given the set of early origin meta-peaks spanning all cell lines, could we accurately classify cell line-specific early origins based on the chromatin landscape and, furthermore, could we also predict the change in relative strength or activity of early origins between cell lines?
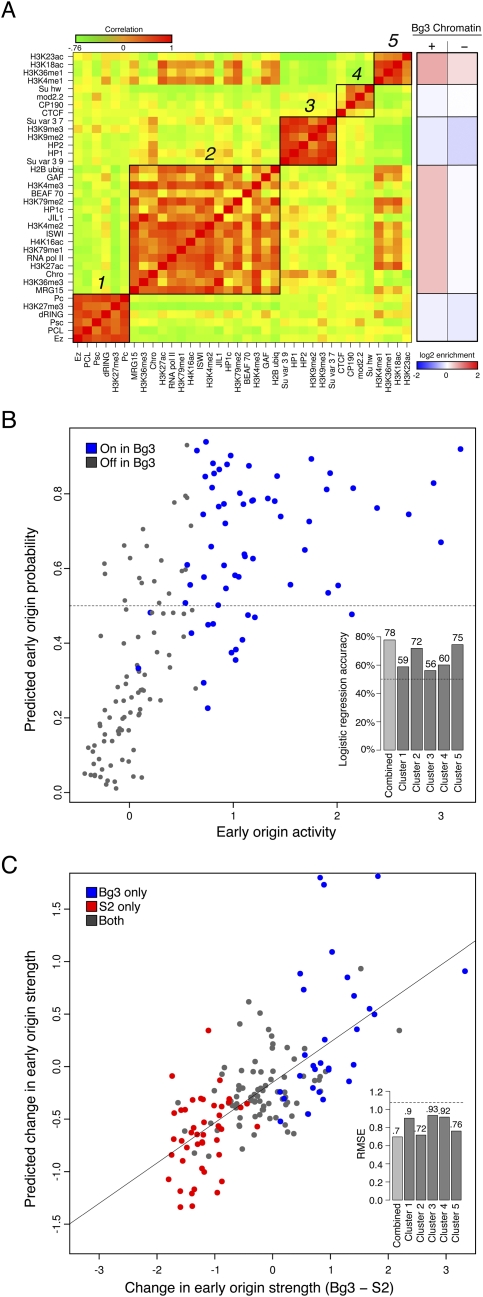
To simplify our description of the chromatin environment at early origins, we sought to identify representative classes of chromatin modifications and DNA-binding proteins that were highly correlated within early activating origins of replication. We first constructed a correlation matrix between each of the normalized chromatin factors and DNA-binding proteins from Bg3 cells, and using hierarchical clustering (Supplemental Fig. S8) we identified five clusters of highly correlated chromatin marks (Fig. 4A, green-red heatmap). Two of these clusters contained predominantly “activating” marks, nucleosome remodelers such as ISWI, and DNA-binding proteins such as RNA Pol II (clusters 2 and 5). In contrast, clusters 1 and 3 were largely composed of repressive chromatin marks and heterochromatin-binding proteins. Finally, insulator elements were also clustered together (cluster 4). These clusters describe the general trends of co-occurrence of chromatin marks and binding factors at the subset of the genome covered by potential early origins. Given the correlations between chromatin marks within each cluster and the redundancy of the “histone code” (Jenuwein and Allis 2001), we reduced the complexity of the 39 modifications and DNA-binding proteins to the integrated signal for each cluster. Specifically, we collapsed the feature vector of marks for each early origin meta-peak down to their mean signal across each of the clusters. A summary of the log2 enrichments for each cluster as a function of origin activity in Bg3 cells is presented in Figure 4A (red-blue heatmap). Of the individual clusters, the mean signals of 2 and 5 correlated highly with early origin strength in Bg3 cells (Pearson's r = 0.43 and 0.56, respectively, Supplemental Fig. S9), while the other clusters showed only weak correlations.
Figure 4.
Chromatin signatures are predictive of early origin activity. (A) Subsets of factors are highly correlated at early origins. (Left heatmap) The pairwise correlation between every factor based on their mean signal at early origin meta-peaks in Bg3 cells was computed (where green indicates a negative correlation and red indicates a positive correlation, with values ranging from −0.76 to 1). Five groups of correlated marks were identified by hierarchical clustering. (Right heatmap) The mean enrichment of each cluster in Bg3 active (+) and Bg3 inactive (−) early origin meta-peaks. (B) Classification of Bg3 early origin usage from the full set of early origin meta-peaks by logistic regression. A logistic regression model using the average chromatin scores of each of the five clusters in Bg3 cells is able to classify (above and below the 0.5 horizontal dashed line as true and false, respectively) with 78% accuracy those meta-peaks that are used in Bg3 on chromosome 3R (blue) and those that are not (gray). (Inset) Predictive power for each cluster individually and the ensemble model. (C) Predicting relative origin strength between Bg3 and S2 cells by linear regression. A linear regression using the change in strength of the chromatin signal from five clusters between Bg3 and S2 is able to predict the change in strength of the early origin meta-peaks between the two cell lines. Predicted change in early origin strength between Bg3 and S2 is plotted as function of actual change. Early origins active in S2 (red) or Bg3 (blue) are indicated. The Pearson correlation is ∼0.7. (Inset) The RMSE over random (horizontal dashed line) for each cluster individually and the ensemble model.
To determine whether the local chromatin environment of the early origin meta-peaks contains enough information to specify which will be active in a given cell line, we built a logistic regression model based on the chromatin clusters to classify the meta-peaks into Bg3-active and Bg3-inactive early origins. To avoid the unique and distinct chromatin signatures of the fourth and the X chromosomes (heterochromatin and hyperacetylation of H4K16, respectively) we restricted our analysis to chromosomes 2L, 2R, 3L, and 3R. Of the 594 early origin meta-peaks on these chromosomes, only 255 were utilized in Bg3 cells (Bg3-active), and the remainder (339) were specific to S2 or Kc cells (Bg3-inactive). Logistic regression allowed the classification of early origin meta-peaks into Bg3-active and Bg3-inactive classes based on a set of predictor variables (the mean cluster strength per meta-peak). We trained our logistic regression model on chromosomes 2L, 2R, and 3L, while holding 3R back for testing. Initially, we trained our model using one chromatin cluster at a time, and found that, unsurprisingly, while all of the clusters were able to predict early origin usage more accurately than a random classifier (Fig. 4B, inset, dashed line), clusters 2 and 5 on their own had the highest accuracy in predicting which of the early origins would be utilized on 3R (Fig. 4B, inset). However, considering all of the clusters in a single logistic regression yielded a classifier with the highest accuracy of all (∼78%; P ≤ 2 × 10−16). We plotted the mean early origin score against the predicted probability of an early origin being Bg3 active, with the actual Bg3-active classifications shown in blue (Fig. 4B).
We next investigated whether we could predict the changes in early origin strength, as determined by mean BrdU incorporation, at early origin meta-peaks between cell lines based solely on chromatin information. We collected chromatin information for the early origin meta-peaks as above for both Bg3 and S2. We then assigned each meta-peak a feature vector containing the change in mean microarray signal for each chromatin cluster between Bg3 and S2. Each meta-peak was also assigned a response variable based on the change in early origin strength between Bg3 and S2 cells. A linear regression model was applied to these data enabling the prediction of change in early origin strength by the change in chromatin cluster strength.
As in the logistic regression above, we trained on 2L, 2R, and 3L, while testing on 3R. We first built initial models using each cluster singly as the input to gauge their individual predictive power. We found that each cluster on its own was capable of outperforming random predictions based on a reduction in root mean squared error (RMSE). However, not surprisingly, we found that changes in clusters 2 and 5 were the most predictive of changes in early origin strength (Fig. 4C, inset). The full linear regression model, taking into account all five of the cluster scores for each early origin, reported the greatest reduction in RMSE, reducing it to ∼0.69. The Pearson correlation between the actual and predicted change in early origin strength for the full model was r ≈ 0.7. The actual change in chromosome 3R early origin strength was plotted against the predicted change with the cell line-specific origins represented by the color of the points (red: active in S2 only, blue: active in Bg3 only) (Fig. 4C). Thus, differences in the local chromatin environment between the two cell lines could account for changes in relative origin activity.
Discussion
Although much progress has been made in understanding how the structure and organization of chromatin regulates the transcription program, we know very little about how start sites of DNA replication are selected and regulated in the context of chromatin. Here, we have used multiple modENCODE data sets to characterize the Drosophila replication program in the context of the surrounding chromatin environment. The computational integration of these diverse data types across multiple cell lines has revealed new insights into how the chromatin landscape influences the selection and regulation of replication origins. Importantly, these insights have allowed us to generate accurate predictive models on the regulation of specific replication origins between cell types.
Potential origins of replication are established in highly dynamic and accessible regions of the genome. We have previously shown that Drosophila ORC-binding sites are surrounded by actively transcribed genes, enriched for the histone variant H3.3 and depleted for bulk nucleosomes (MacAlpine et al. 2010). Underscoring the importance of nucleosome occupancy in the selection of replication origins, recent studies in S. cerevisiae have shown that nucleosome occupancy is a determinant for ORC binding and that the precise ORC-dependent positioning of nucleosomes flanking the origin is critical for origin function (Berbenetz et al. 2010; Eaton et al. 2010). In both yeast and Drosophila, the nucleosomes surrounding potential origins of replication marked by ORC are dynamic and undergo rapid nucleosome exchange (Kaplan et al. 2008; Deal et al. 2010). This chromatin turnover may involve specific chromatin remodeling activities.
Consistent with the idea of chromatin remodelers altering chromatin organization at origins of replication, we found that Drosophila ORC-binding sites are highly enriched for the ATP-dependent chromatin remodeler ISWI. ISWI can act as part of the NURF complex, specific subunits of which were also enriched (NURF301) (Längst and Becker 2001). The chromatin-binding proteins WDS and GAF were also enriched at ORC-binding sites and have both been implicated in facilitating nucleosome dynamics (Petesch and Lis 2008; Suganuma et al. 2008). Finally, DNA-binding proteins with chromatin remodeling activities or functions were among the most discriminatory features for ORC binding in our SVM analysis. We propose that active chromatin dynamics facilitated by remodeling activities will be a conserved feature of replication origins in all eukaryotes. In support of this hypothesis, mammalian replication origins are also enriched in the vicinity of active promoters (Cadoret et al. 2008; Sequeira-Mendes et al. 2009; Karnani et al. 2010).
In S. cerevisiae, the precise nucleosome positioning observed at origins of replication can be reconstituted in vitro with purified ORC, recombinant histones, and ISWI (Eaton et al. 2010). At this point, we do not know whether ORC is directly interacting with remodeling enzymes and recruiting them to origins of replication or, alternatively, if ORC is being recruited to dynamic and open chromatin facilitated by the chromatin remodeling activity. Future experiments will address these questions.
Despite the simplicity and mostly invariant nature of the underlying DNA code, the transcription program is able to respond to almost limitless developmental and environmental perturbations. It is the complex interactions between regulatory networks of chromatin modifications, transcription factors, nucleosome positioning, and DNA accessibility that provide for the remarkable plasticity in gene expression derived from the genome. It is becoming increasingly clear that the replication program responds to many of the same epigenetic cues that regulate the transcription program. For example, X chromosome dosage compensation in the fly results not only in the H4K16ac-dependent transcriptional up-regulation of X-specific genes in male cells (Laverty et al. 2010), but also a sex chromosome-specific change in replication timing (Schwaiger et al. 2009). Similarly, we find that ORC, early origins, and early replicating regions of the genome are highly enriched for activating chromatin marks (H3K4me, H3K18ac, H3K27ac, etc.). This coordination between the transcription and replication programs is likely critical for the expression and inheritance of genetic and epigenetic information.
The integration of multiple modENCODE data sets across three different Drosophila cell lines has allowed us to generate predictive models of ORC binding, origin usage, and origin strength. Sequence, chromatin modifications, and DNA-binding proteins all contribute in an additive manner to our ability to identify ORC-binding sites in the genome. Additionally, our data suggest that the competency of a genomic location to replicate in the presence of HU (i.e., to act as an early origin of replication) is determined by the local chromatin landscape. By integrating the chromatin signals near early activating origins of replication, we were able to use logistic regression models to classify with a high degree of accuracy (∼78%) which potential early origins would be utilized within a cell line. Not only were we able to predict origin utilization, but the same chromatin marks were also used to build linear regression models that could predict the relative strength or activity of early origins between two cell lines. Our ability to predict the strength of origin usage between cell lines based on the surrounding chromatin environment suggests that the chromatin environment acts as a rheostat and not a binary switch. That is, the signals from multiple activating and repressive chromatin marks are assimilated to provide a relative index of the potential for a sequence to function as an origin of replication. Finally, although we describe the replication program as responding to the local chromatin environment, there is increasing evidence that the replication program is also important for epigenetic memory (Zhang et al. 2002; Lande-Diner et al. 2009).
Methods
Cell growth
Kc167 and S2-DRSC cells were cultured in 150-mm plates in Schneider's Insect Cell Medium (Invitrogen) supplemented with 10% FBS and 1% penicillin/streptomycin/glutamine (Invitrogen). ML-DmBG3-c2 cells were cultured as above with 10 μg/mL human insulin (Sigma). All cell growth was conducted at 25°C. Cell cycle position was determined by flow cytometry.
ChIP-seq
Chromatin immunoprecipitations were performed as in MacAlpine et al. (2010) using a polyclonal ORC2 antibody (Austin et al. 1999). Sequencing libraries were generated using the ChIP-Seq sample prep kit and protocol (Illumina, http://grcf.jhmi.edu/hts/protocols/). Libraries were sequenced on a GAII Illumina sequencer and processed using SCS2.6 software.
Read mapping and peak calling
MAQ (Li et al. 2008) was used to map the reads back to release 5 of the D. melanogaster genome. Reads with a quality score ≥35 were considered in the subsequent analysis to filter out the reads that could not be mapped uniquely. ORC ChIP-seq peaks were called using PeakSeq (Rozowsky et al. 2009) with default parameters. Input sequencing libraries were used to control for sequencing specific biases. Replicates were combined by intersection; overlapping peak calls were considered verified and were reduced in a per-nucleotide union. Replicates passed quality control if 80% of the top 40% of peaks (by strength) in one experiment existed in the other and vice-versa. Whole-genome background-subtracted density tracks were produced by the R package SPP (Kharchenko et al. 2008).
Replication timing
Approximately 2.7 × 108 Kc167, S2-DRSC, or ML-DmBg3-C2 cells were treated with HU to a final concentration of 1 mM and allowed to incubate for 3 h. BrdU was then added to 50 μg/mL final concentration to half the cells, and both cell populations were incubated for an additional 21 h. Cells treated only with HU are the late timing samples, while cells treated with HU and BrdU are considered the early timing samples. All cells were then washed one time with cold 1× PBS and replated. Early timing cells were treated with an additional 50 μg/mL BrdU for 1 h and then harvested for DNA extraction. The late timing cells were incubated for 4 h after washing, followed by a 2-h incubation with 50 μg/mL BrdU before being harvested for genomic DNA extraction.
Early origin mapping
Approximately 3.5 × 108 Kc167, S2-DRSC, or ML-DmBg3-C2 cells were treated with hydroxyurea (HU) (Sigma) to a final concentration of 1 mM and incubated for 3 h. 5-bromo-2-deoxyuridine (BrdU) (Roche) was then added to a final concentration of 50 μg/mL for 18–20 h. Cells treated solely with HU served as the control sample. Cells were harvested, centrifuged at 1000_g_ for 5 min, and washed once with cold 1× PBS, and the resulting pellets were used for genomic DNA extraction.
DNA extraction
For early origin experiments, pellets were resuspended in 2 mL of 10 mM Tris (pH 9.5) and 2 mL of NDS (10 mM Tris at pH 9.5, 500 mM EDTA at pH 9.5, 1% SDS); after addition of 0.5 mL of 20 mg/mL proteinase K, samples were inverted to mix and incubated at 37°C for 2 h. Samples were phenol/chloroform extracted twice and allowed to precipitate overnight at 25°C. DNA was pelleted at 3500 rpm for 12 min, washed in 70% ethanol, and resuspended in 4 mL of 1× TE. A total of 20 μL of 10 mg/mL RNaseA was added to samples and allowed to incubate at 37°C for 1–2 h. DNA was precipitated with 3 M NaOAc and cold ethanol and resuspended in 500 μL of 1× TE. DNA was sheared to an average size of 1 kb. For timing experiments, DNA was extracted as above, but in the following volumes: 0.7 mL of 10 mM Tris (pH 9.5), 0.7 mL of NDS, 0.1 mL of 20 mg/mL proteinase K. DNA was resuspended in 250 μL of 1× TE.
BrdU immunoprecipitation
A total of 50 μL of Dynabeads M-280 sheep anti-mouse IgG beads (Invitrogen) per sample were washed three times in 1 mL of cold 1× PBS/5 mg/mL BSA using a magnetic concentrator and resuspended in the same. Next, 5 μL of 0.5 mg/mL anti-BrdU antibody (Roche) was added and allowed to incubate with rotation overnight at 4°C. Beads were washed as above and resuspended in 50 μL of the same; 15 μg of DNA was added to new tubes, incubated at 100°C for 5 min, and cooled on ice. Then, 450 μL of RIPA (50 mM Hepes-KOH at pH 7.6, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% Na-deoxycholate) was added to the DNA with the prepped beads and allowed to incubate overnight with rotation at 4°C. Beads were collected using a magnetic concentrator and the supernatant saved as the INPUT. Beads were washed four times in 1 mL of cold RIPA and one time in 1 mL of 1× TE, with 5-min room temperature rotations between washes. Beads were resuspended in 150 μL of TE/1% SDS and incubated at 65°C for 10 min with two to three quick vortexes. Supernatants were saved, to which 150 μL of TE, 300 ng/L glycogen, and 1 μg/μL proteinase K were added, and incubated for 1 h at 37°C. A total of 12 μL of 5 M NaCl was added to the samples and subsequently phenol/chloroform extracted. DNA was resuspended in 15 μL of dH2O.
Array hybridization and analysis
Labeling of DNA was performed as in MacAlpine et al. (2010). All experiments were performed using biological triplicates. Labeled DNA was hybridized on custom whole-genome, 244K tiling microarrays (Agilent). Slides were hybridized and washed as per Agilent recommendations. The array data was processed and analyzed as previously described (MacAlpine et al. 2010).
The _P_-value cited for the number of early origin peaks overlapping an ORC peak was derived by generating R = 100,000 sets of random segments that mirrored the early origin peaks in width, number, and chromosome membership, and by counting how many of those segments overlapped an ORC peak. We then found n, the number of samples in this bootstrap distribution that had greater than or equal to the number of ORC peaks overlapped in the early origin meta-peak set. Then, P = (n + 1)/(R + 1).
Meta-peaks
To construct a set of regions in the genome with the potential to bind ORC or to host early origin activity, we constructed a set of ORC meta-peaks and early origin meta-peaks, respectively. These meta-peaks are contiguous nucleotides that were covered by a peak in at least one cell line. To conservatively account for the possibility of one peak in a particular cell line overlapping multiple peaks in another cell line, we built the Venn diagram using the set of ORC meta-peaks, such that multiple peaks in a meta-peak were scored as a single overlap.
SVM analysis
SVM analysis was performed on the set of annotated ORC-binding sites for each cell line using the LIBSVM suite (Chang and Lin 2001). The SVM was trained and tested on a positive and negative sequence set. The former consisted of 500-nt sequences centered on the ORC-binding sites. The latter (random set) contained an equal number of 500-nt sequences selected randomly from the genome, with the condition that they exclude the positive set and that the chromosome frequency and proportion of promoter proximal instances match that of its positive counterpart. Three feature sets were used: sequence 1–6 mers, chromatin marks, and protein-binding sites. The _k_-mer feature values were compiled as frequencies of each _k_-mer in each 500-nt sequence (counting each _k_-mer and its reverse complement as the same feature), then scaled relative to the rest of the sequence set to the range 0 ≤ x ≤ 1. Both the chromatin marks and protein-binding sites were given binary values 0.1 depending on whether they fell on each 500-nt sequence. The entire feature set was then split into a training set consisting of sequences on chromosomes 2L, 3L, and 3R, and a testing set containing sequences on chromosome 2R. The SVM was then trained on the training set with 10-fold cross validation. Testing was performed on 2R, and the performance of the classifier was evaluated based on the resulting ROC curve. This analysis was carried out using each of the three feature types individually, as well as all of the features simultaneously. In addition, the discriminative power of each individual feature was determined by ranking them based on their _F_-score and their predilection toward one class over another determined by the class proximity, defined here as _t_-statistic obtained from a Student's _t_-test comparing feature counts in the positive and negative sets.
Motif analysis
Motifs were discovered in the 500-nt sequences containing the ORC-binding sites via MEME (Bailey and Elkan 1994), then ranked according to their _P_-value. The top three motifs were then recovered from the whole-genome using MAST (Bailey and Gribskov 1998). The motif loci delivered by MAST were then ranked by _P_-value, and those that fell on the ORC-binding site-containing sequences were marked. The presence of these motifs was correlated to ORC-binding sites by generating a ROC curve denoting the false and true positive rate of their appearance in the ORC set over random genomic loci.
Chromatin enrichment heatmaps
The list of 39 factors from the Karpen group (Kharchenko et al. 2011) includes the following: EZ, MRG15, H3K36me3, SU(VAR)3-9, PCL, CHRO, H3K27ac, PSC, H3K79me1, H3K9ac, RNA Pol II, H3K27me3, H4K16ac, CTCF, ISWI, H3K4me2, JIL1, HP1C, HP1, WDS, H3K4me1, H3K36me1, HP2, H3K9me2, dRING, CP190, H3K4me3, H3K79me2, BEAF-70, H3K9me3, H3K18ac, GAF, H3K23ac, MOD2.2, NURF301, SU(HW), PC, H2B-ubiq, and SU(VAR)3-7. We also used cell-line-specific RNA-seq data from the Celniker group (Graveley et al. 2011) and the 20-min CATCH-IT data, H2Av, H3.3, and nucleosome density data from the Henikoff group in S2 cells (Henikoff et al. 2009; Deal et al. 2010).
ORC and early origin heatmaps
Each factor available for comparison (nucleosome dynamics, RNA-seq, chromatin binders, and histone marks) was quantile normalized per-probe or per-transcript between cell lines. Each factor was already mean shifted and loess smoothed appropriately by its originating group. Each factor's distribution was then scaled to unit variance, and the median of the scores in 1-kb windows centered on ORC peaks or directly under early origin peaks was calculated for the ORC matrix (Fig. 2C) and the early origin matrix (Fig. 2B), respectively.
Replication timing heatmap
To generate the replication timing correlations for the nucleosome dynamics, chromatin mark, and factor data, each replication timing probe was paired with a collection of factor probes that it overlapped. These factor probes were averaged per timing probe, and a Spearman's ρ correlation was taken between the timing probe values and the mean factor probe values. For the RNA-seq data, each transcript was assigned a timing score based on the median probe value of the timing probes that it overlapped, and a Spearman's ρ was taken of these paired samples.
Regression analysis
The early origin meta-peak set was created by taking a per-nucleotide union of the three cell lines' early origin peak sets and then combining contiguous nucleotides into meta-peaks. This yielded a set of 823 “early origin meta-peaks.” These early origin meta-peaks were given a score for each cell line corresponding to the mean early origin microarray signal within the meta-peak. Each meta-peak was also given a vector of scores corresponding to the microarray signal for each of the 36 factors produced by the Karpen group (Kharchenko et al. 2011). In selecting factors from the Karpen group, we limited ourselves to those that were common between Bg3 and S2. The list of factors was as follows: EZ, MRG15, H3K36me3, SU(VAR)3-9, PCL, CHRO, H3K27ac, PSC, RNA Pol II, H3K79me1, H4K16ac, CTCF, ISWI, H3K4me2, JIL1, HP1C, HP1, H3K4me1, H3K36me1, HP2, H3K9me2, DRING, CP190, H3K27me3, H3K79me2, BEAF-70, H3K4me3, H3K9me3, H3K18ac, GAF, H3K23ac, MOD2.2, SU(HW), PC, H2B-ubiq, and SU(VAR)3-7. Every factor was quantile normalized between cell lines and then mean centered and divided by the standard deviation (_Z_-score transformation). To produce the correlation heatmap in Figure 4A, a pairwise correlation matrix was constructed using Pearson's correlation (r) and then clustered using (1-[_r_]) as the distance matrix with Ward's method for hierarchical clustering (Ward 1963). The enrichment heatmap in Figure 4A represents the mean enrichment for all Bg3 factors in a cluster within Bg3 active (+) and Bg3 inactive (−) early origins.
Logistic regression
The early origin meta-peak set was split into a training set (chromosomes 2L, 2R, and 3L; 441 meta early origins) and a test set (3R; 153 meta early origins). Each early origin meta-peak was given a set of five predictors corresponding to the mean of the factors within each of the five clusters. Each early origin meta-peak was also given a logical response variable, which was true when the early origin meta-peak was active in Bg3 (i.e., if the early origin meta-peak overlapped a called early origin peak from Bg3) and false if it was inactive in Bg3. We then used logistic regression to regress from the mean cluster scores to the Bg3-active response variable using the glm function of R (R Development Core Team 2008) with a binomial logit link function. The model parameters were fit using 100-fold cross validation. This regression was then put through a stepwise model selection process minimizing the Akaike information criterion (AIC), during which clusters 1, 2, and 5 were selected for the final model. Accuracy was gauged on 3R as (True Positives + True Negatives)/n, where n was the total number of early origin meta-peaks.
Linear regression
The early origin meta-peak set was split into training and test sets as above. The response variable in this case was the difference in mean microarray signal within each early origin meta-peak between Bg3 and S2. Likewise, the five predictor variables took the form of the difference in mean signal strength between each of the five clusters between Bg3 and S2. We then used standard linear regression using all five clusters with parameters again fit by 100-fold cross validation to predict the difference in early origin strength via the difference in cluster strengths. Accuracy was judged by a reduction in RMSE and compared with a background RMSE determined by the mean RMSE of 1000 random permutations of the pairing between the predicted change in early origin strength and the actual change in early origin strength.
Data accession
All data has been deposited at GEO. GSE17281, ML-DmBG3-c2 Replication Timing; GSE17279, Kc167 Replication Timing; GSE17280, S2-DSRC Replication Timing; GSE17287, ML-DmBG3-c2 Replication Origins; GSE17285, Kc167 Replication Origins; GSE17286 S2-DRSC, Replication Origins; GSE20888, ORC2 ML-DmBG3-c2 ChIP-Seq; GSE20889, ORC2 KC-167 ChIP-Seq; GSE20887, ORC2 S2-DSRC ChIP-Seq.
Acknowledgments
We thank Alexander Hartemink for his discussion and advice and the members of the modENCODE consortium for ideas and proposed analyses. This work was supported by the Whitehead Foundation Scholar Award (D.M.M.) and the National Institutes of Health grant HG004279 (D.M.M.).
Footnotes
References
- Aggarwal BD, Calvi BR 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376 [DOI] [PubMed] [Google Scholar]
- Austin R, Orr-Weaver T, Bell S 1999. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev 13: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bailey TL, Gribskov M 1998. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 14: 48–54 [DOI] [PubMed] [Google Scholar]
- Bell S, Dutta A 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Berbenetz NM, Nislow C, Brown GW 2010. Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet 6. doi: 10.1371/journal.pgen.1001092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier A, Chatterji S, Cozzarelli N 2004. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol 5: R22 http://genomebiology.com/2004/5/4/R22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN 2008. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-C, Lin C-J 2001. LIBSVM: A library for support vector machines. Software available at http://www.csie.ntu.edu.tw/cjlin/libsvm
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Méchali M 2004. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol 6: 721–730 [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S 2010. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328: 1161–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM 2010. Conserved nucleosome positioning defines replication origins. Genes Dev 24: 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM 2010. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet 11: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Tabib A, Hecht M, Cedar H 2008. DNA replication timing of the human β-globin domain is controlled by histone modification at the origin. Genes Dev 22: 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin J, Yang L, Artieri C, van Baren MJ, Booth BW, Brown JB, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature (in press). doi: 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K 2009. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res 19: 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD 2001. Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Fried-man N, Rando OJ 2008. Cell cycle-and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet 4: e1000270 doi: 10.1371/journal.pgen.1000270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor C, Malhotra A, Dutta A 2007. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res 17: 865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A 2010. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell 21: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle N, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature (in press). doi: 10.1038/nature09725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Viggiani CJ, Tavare S, Aparicio OM 2009. Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev 23: 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande-Diner L, Zhang J, Cedar H 2009. Shifts in replication timing actively affect histone acetylation during nucleosome reassembly. Mol Cell 34: 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längst G, Becker PB 2001. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J Cell Sci 114: 2561–2568 [DOI] [PubMed] [Google Scholar]
- Laverty C, Lucci J, Akhtar A 2010. The MSL complex: X chromosome and beyond. Curr Opin Genet Dev 20: 171–178 [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford J, Bell S 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell 7: 21–30 [DOI] [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B 2005. Chromatin remodeling in dosage compensation. Annu Rev Genet 39: 615–651 [DOI] [PubMed] [Google Scholar]
- MacAlpine D, Rodriguez H, Bell S 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18: 3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255: 817–823 [DOI] [PubMed] [Google Scholar]
- Miotto B, Struhl K 2010. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by geminin. Mol Cell 37: 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The modENCODE Consortium 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak D, Pflumm M, Chesnokov I, Huang D, Kellum R, Marr J, Romanowski P, Botchan M 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91: 311–323 [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at HSP70 loci. Cell 134: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2008. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Rando OJ, Chang HY 2009. Genome-wide views of chromatin structure. Annu Rev Biochem 78: 245–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beall E, Botchan M 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J 23: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Shaffer CD, Elgin SCR 2009. A lot about a little dot - lessons learned from Drosophila melanogaster chromosome 4. Biochem Cell Biol 87: 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB 2009. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol 27: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley J 1998. A Mec1-and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schubeler D 2009. Chromatin state marks cell-type-and gender-specific replication of the Drosophila genome. Genes Dev 23: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M 2009. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5: e1000446 doi: 10.1371/journal.pgen.10001092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621 [DOI] [PubMed] [Google Scholar]
- Simpson RT 1990. Nucleosome positioning can affect the function of a _cis_-acting DNA element in vivo. Nature 343: 387–389 [DOI] [PubMed] [Google Scholar]
- Suganuma T, Gutiérrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL 2008. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol 15: 364–372 [DOI] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly T, Walter J 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev 17: 1894–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer B, Grunstein M 2002. Histone acetylation regulates the time of replication origin firing. Mol Cell 10: 1223–1233 [DOI] [PubMed] [Google Scholar]
- Ward JH Jr 1963. Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58: 236–244 [Google Scholar]
- Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H 2002. Establishment of transcriptional competence in early and late S phase. Nature 420: 198–202 [DOI] [PubMed] [Google Scholar]