The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer (original) (raw)
Abstract
The heterochromatin protein 1 (HP1) family of proteins is involved in gene silencing via the formation of heterochromatic structures. They are composed of two related domains: an N–terminal chromo domain and a C–terminal shadow chromo domain. Present results suggest that chromo domains may function as protein interaction motifs, bringing together different proteins in multi-protein complexes and locating them in heterochromatin. We have previously determined the structure of the chromo domain from the mouse HP1β protein, MOD1. We show here that, in contrast to the chromo domain, the shadow chromo domain is a homodimer. The intact HP1β protein is also dimeric, where the interaction is mediated by the shadow chromo domain, with the chromo domains moving independently of each other at the end of flexible linkers. Mapping studies, with fragments of the CAF1 and TIF1β proteins, show that an intact, dimeric, shadow chromo domain structure is required for complex formation.
Keywords: chromatin structure/chromo domain/heterochromatin protein 1/protein complex/protein structure
Introduction
The first heterochromatin-associated protein to be characterized was heterochromatin protein 1 (HP1), a suppressor of position effect variegation (PEV) (James and Elgin, 1986; Eissenberg et al., 1990). HP1 is found in complexes with other known heterochromatin proteins, e.g. Su(var)3–7 (Cleard et al., 1997) and Su(var)3–9 (Aagaard et al., 1999), and its mutation causes recessive embryonic lethality due to defects in chromosome morphology, lengthened prophase and subsequent chromosome segregation (Kellum and Alberts, 1995). Mutations can also result in aberrant association of chromosomes and multiple telomeric fusions (Fanti et al., 1998).
HP1 homologues have been found in many other organisms from Schizosaccharomyces pombe (Lorentz et al., 1994; Ekwall et al., 1995) to mammals (Singh et al., 1991; Saunders et al., 1993). There are three HP1 protein family members in mammals, HP1α, HP1β (MOD1) and HP1γ (MOD2), and different patterns of phosphorylation and localization point to possible differences in their function (Minc et al., 1999).
In Drosophila, phosphorylation of HP1 is required for efficient heterochromatin targeting and, possibly, heterochromatin assembly (Eissenberg et al., 1994; Zhao and Eissenberg, 1999). In mammals, HP1α and HP1γ exist in different phosphorylated forms, becoming hyper-phosphorylated at mitosis. In contrast, HP1β remains as a unique isoform throughout the cell cycle (Minc et al., 1999). In Drosophila, HP1 is localized to heterochromatin, to telomeres and to discrete regions of euchromatin (Kellum et al., 1995; Fanti et al., 1998). In mouse and human cells, HP1α is found predominantly in centromeres, HP1β (MOD1) is distributed widely on the chromosome, and HP1γ (MOD2) localizes mostly to euchromatin (Minc et al., 1999); their localization changes during the cell cycle in both Drosophila (Kellum et al., 1995) and mammals (Minc et al., 1999; Murzina et al., 1999). Recently, we have shown that as mitosis is approached some HP1β dissociates from heterochromatin, as histone H3 becomes hyper-phosphorylated, and then reassociates at the end of mitosis when H3 is dephosphorylated (Murzina et al., 1999).
The HP1 proteins make up one class of chromo domain proteins (Paro and Hogness, 1991), having an N–terminal chromo domain and a related C–terminal shadow chromo domain (Aasland and Stewart, 1995; Koonin et al., 1995). Many other chromo domain proteins are also involved in the regulation of gene expression resulting from alterations in chromatin structure (Cavalli and Paro, 1998). The polycomb protein (Pc) represents a second important class, in which an N–terminal chromo domain is present, but the shadow domain is not. These generally much larger proteins contain a different conserved sequence motif in the C–terminus (Paro, 1990; Paro and Hogness, 1991). The construction of chimeric proteins, consisting of either the HP1 protein with its chromo domain replaced by that from Pc, or the Pc protein with an HP1 chromo domain, has shown that both the chromo and shadow domains are important for correct chromatin localization. These results suggested that they may interact independently with different targets in heterochromatin (Platero et al., 1995).
Although HP1 proteins are located in chromatin, they do not appear to bind to DNA directly (Singh et al., 1991; Ball et al., 1997). Rather, they have been found to interact with a number of different proteins. So far, the only known example of an interaction involving the chromo domain is the interaction of Drosophila HP1 with the origin recognition complex (ORC) that is required for initiation of eukaryotic DNA replication (Pak et al., 1997). All other known interactions are mediated via the shadow chromo domain. The importance of the shadow chromo domain in HP1 function is emphasized by the fact that a truncated HP1 mutant Su(var)2–504, lacking part of the shadow domain, does not localize in either heterochromatin, euchromatin or at telomeres (Fanti et al., 1998).
Both the mouse and human HP1 proteins have been shown to interact with the transcriptional intermediary factors (TIFs) α and β (Le Douarin et al., 1996; Nielsen et al., 1999; Ryan et al., 1999). TIF1β (or KAP1) also binds to proteins containing the KRAB domain (Kruppel-associated box), one of the most widely distributed transcriptional repressor domains in mammals (Friedman et al., 1996; Moosmann et al., 1996). It has been suggested that the HP1–TIF–KRAB complex might recruit hetero– chromatin-like complexes to specific loci on the chromosome defined by the DNA binding site of the Kruppel transcription factors (Ryan et al., 1999). TIF1α is a nuclear protein that interacts directly with the ligand-dependent activation domain of certain nuclear hormone receptors to suppress transcription (Le Douarin et al., 1995); it has also been shown to be located in euchromatin (Remboutsika et al., 1999). Both TIF1α and TIFβ appear to repress transcription via histone deacetylases (Nielsen et al., 1999). Recently, we have demonstrated that the large subunit of chromatin assembly factor 1 (CAF1p150) binds to mouse HP1 proteins. The interaction is required for the association of CAF1 with heterochromatin in non-S-phase cells, and CAF1 also promotes the incorporation of MOD1 into nascent chromatin during DNA replication in vitro. We identified a peptide motif that is conserved in both the TIFs (Le Douarin et al., 1996) and the large subunit of CAF1p150 that interacts with the shadow chromo domain (Murzina et al., 1999).
Thus the results so far suggest that the HP1 proteins may function as adaptors, bringing together different proteins in multi-protein complexes, and locating them in heterochromatin via protein–protein interactions with the chromo and shadow chromo domains. In order to understand the mechanisms involved we are studying the structure and interactions of the mouse HP1β protein, MOD1 (Murzina et al., 1999). We show here that the full-length MOD1 protein is a dimer where the interaction is mediated via the C–terminal shadow chromo domain (MOD1C). We present the structure of MOD1C and show that an intact dimer structure is required for the interaction with the CAF1 and TIF1β proteins.
Results
Shadow domain structure
Based on limited proteolysis data (Ball et al., 1997), and a sequence alignment of the different HP1 proteins (Figure 1), residues 104–171 of MOD1 were expressed in Escherichia coli and purified for structural studies. The purified MOD1C protein had the expected amino acid composition and molecular mass (data not shown). Studies of 15N relaxation of the backbone amides suggested that the protein was larger and tumbled more slowly in solution than the homologous chromo domain (Figure 2). Equilibrium sedimentation analysis subsequently confirmed that the shadow chromo domain is dimeric in solution (data not shown).
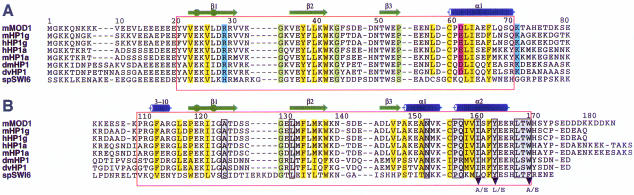
Fig. 1. Sequences of the chromo domains (A) and the shadow chromo domains (B) from different HP1 proteins, numbered so that they correspond to MOD1. Secondary structure elements observed in MOD1 are shown above the alignments; cylinders represent 310 (3–10) or α–helices (α1 and α2), arrows represent β–strands and circles indicate β–bulges. For each domain the residues that make up the hydrophobic core of a ‘subunit’ are shaded in yellow and other residues considered important for the structure are shown in green (Gly and Pro). Charged residues in the chromo domain, which are replaced by hydrophobic residues in the shadow chromo domain, are coloured blue (basic) and red (acidic). The red boxes enclose the structured parts of the proteins. Residues that form the dimer interface in the shadow chromo domain are boxed and shaded in grey. Mutations described in this paper are indicated below the alignment. The proteins are, from the top, mouse MOD1/human HP1β (residues 1–81 and 103–185), mouse HP1γ (1–80 and 97–173), human HP1γ (1–80 and 97–173), human HP1α (1–80 and 106–191), mouse HP1α (1–80 and 106–191), Drosophila melanogaster HP1 (1–84 and 132–206), Drosophila virilis HP1 (1–84 and 139–213) and Schizosaccharomyces pombe SWI6 (59–145 and 252–328).
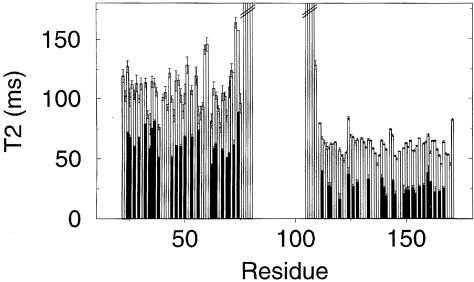
Fig. 2. Backbone 15N T2 relaxation times for full-length MOD1 (black), free N–terminal domain and free C–terminal domain (both white). Amide groups in full-length MOD1 were assigned (where possible) by comparing spectra of the full-length protein with those of the individual domains. The T2s were calculated by non-linear least-squares fitting (Broadhurst et al., 1995).
The structure of MOD1C is well defined except for the five N–terminal residues, 104–109, and the C–terminal residue, 171, which are flexible in solution as judged by the 15N relaxation experiments (see Figure 3). The shadow chromo domain structure thus corresponds to residues 110–170 of mouse MOD1. The structures satisfy the experimentally derived distance restraints, with on average less than one violation >0.5 Å per structure, and have good stereochemistry and van der Waals packing (see Table I).
Fig. 3. The structure of the shadow chromo domain dimer from MOD1. The backbone r.m.s.d. for the structure is 0.63 Å over the monomer and 0.81 Å over the dimer. (A) A stereo plot of the backbone traces of the ensemble of 16 calculated structures; the two monomers are depicted in red and blue. (B) A cartoon representation of the shadow chromo domain dimer (again red and blue) with the chromo domain from MOD1 (yellow) for comparison. (C) A close up stereo view of the inter-monomer interface with the side chains of interfacial residues shown; key residues are labelled. These plots were produced using MOLSCRIPT and Raster3D (Kraulis, 1991; Merritt and Bacon, 1997).
Table I. Structural statistics for the final ensemble of 16 refined structures of the HP1β shadow chromo domain.
| | All structures | Closest to mean | | | -------------------------------------------------------------------- | ---------------- | ------ | | NOE restraints | | | | no. of restraints used | 2090 | 2090 | | average restraint violation (Å) | 0.0061 ± 0.0006a | 0.0055 | | Coordinate r.m.s.d. (computed over residues 110–170) | | | | backbone atoms | | | | over one monomer (Å) | 0.63 | 0.42 | | over both monomers (Å) | 0.81 | 0.62 | | all heavy atoms | | | | over one monomer (Å) | 1.08 | 0.85 | | over both monomers (Å) | 1.17 | 0.95 | | Parameter r.m.s.d. from idealized geometry | | | | bonds (Å) | 0.002 ± 9×10–5b | 0.002 | | angles (°) | 0.333 ± 0.010b | 0.329 | | improper dihedrals (°) | 0.225 ± 0.018b | 0.201 | | Final energy | | | | _E_L–Jc (kJ/mol) | –441 ± 44 | –360 | | Ramachandran plot quality (location of non-Gly and non-Pro residues) | | | | most favoured region (%) | 69.9 | 69.8 | | additionally allowed region (%) | 21.8 | 22.6 | | disallowed region (%) | 2.8 | 5.7 |
As expected from their homology, each monomer of MOD1C forms a compact fold very similar to that of the chromo domain (see Figure 3). MOD1C, however, forms a symmetrical dimer, burying 687 ± 47 A2 of surface area, in which the interface principally involves the C–terminal α–helices of each monomer. Residues that form the dimer interface in the shadow chromo domain are indicated in Figure 1; the main contacts involve I161, Y164 and L168 (see Figure 3C). There are also significant contacts between residue 153 in the α1 helix of one monomer and residue 161 in the α2 helix of the other, between residue 158 and the peptide backbone of residue 154, and between the side chain of W170 and A125, L132 and Y164.
Mutations disrupting the dimer structure
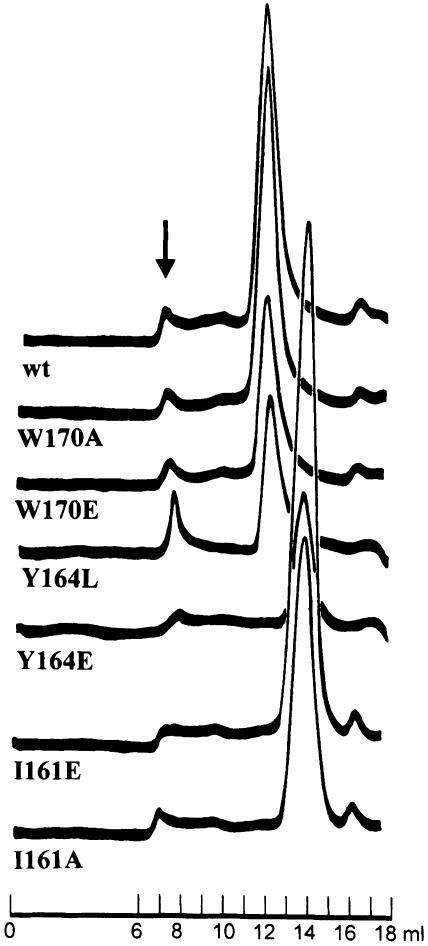
The importance of particular residues for dimer formation was investigated by mutating residues either to those found in the monomeric chromo domains or to glutamate, thereby introducing a negative charge into the hydrophobic dimer interface. We then determined the relative size of the different mutant proteins (I161 to A or E, Y164 to L or E and W170 to A or E) using gel-filtration chromatography. As shown in Figure 4, wild-type MOD1C and the Y164L, W170A and W170E mutants eluted at the same column volume, suggesting that the size and shape of each mutant protein are similar to those of the wild type. Changing Y164 to L does not disrupt dimer formation, but did decrease the stability of the protein, which showed a tendency to aggregate and precipitate from solution. In contrast, the Y164E, I161A and I161E mutants appear to have a lower molecular weight than the wild-type protein (see Figure 4). We confirmed that each of these proteins was intact by SDS–PAGE and that the Y164E and I161E mutants were similar in structure to the wild-type protein by circular dichroism (data not shown). Sedimentation analysis of these mutants showed that the I161E mutant is monomeric, whereas the Y164E mutant showed weak association with a _K_d of ∼500–600 μM (data not shown). Taken together, the data show that mutation of either residue 161 or 164 results in a protein that is structured and essentially monomeric. The results also confirm the importance of these residues for formation of the dimer interface.
Fig. 4. MOD1C mutations disrupting the dimer structure. Wild-type MOD1C and the mutants were expressed as His–tagged fusions in E.coli, purified with Ni–NTA spin columns (Qiagen) and loaded directly onto a Superdex S75 gel–filtration column (24 ml bed volume) to assess the size of the proteins. Gel-filtration elution profiles are presented for wild-type MOD1C (wt) and the mutants W170A, W170E, Y164L, Y164E, I161E and I161A. The arrow indicates aggregates that eluted in the void volume of the column.
The structure of intact MOD1
We have shown previously that the recombinant chromo domain exists as a monomer in solution (Ball et al., 1997), and in this work we have determined that the shadow chromo domain is a dimer. Sedimentation analysis of the intact protein showed that the molecular weight in solution is 42 kDa, approximately twice that calculated from the sequence (21.5 kDa). These results indicate that the full-length protein is also a dimer, but left open the question as to what inter-domain interactions occur in native MOD1.
The mobility of the individual domains can be estimated from the backbone 15N T2 relaxation times, which depend on the rotational correlation time of the molecule—larger and more slowly tumbling molecules having shorter T2 values. The average 15N T2 is 99 ms for the free N–terminal domain and 62 ms for the free C–terminal domain. These data show that the free N–terminal domain tumbles significantly faster in solution than the free C–terminal domain, consistent with the former being a monomer and the latter a dimer. For the full-length protein, the average 15N T2 is 61 ms for the N–terminal domain and 26 ms for the C–terminal domain. (The individual values and the actual residues compared are shown in Figure 2.) In the full-length protein both domains have shortened T2 values, consistent with a larger overall structure. Significantly, however, the T2 values in the C–terminal domain remain lower than those in the N–terminal domain. The difference in 15N T2 values between the two domains suggests strongly that they have different mobility in the full-length protein and must therefore be moving largely independently of each other. The consistently longer T2 values for the N–terminal domain suggest that it remains unassociated with other parts of the molecule in the full-length protein. These conclusions are also supported by the fact that the linker region between the two domains is unstructured (see Figure 2), consistent with its high level of accessibility to proteases (Ball et al., 1997). In summary, the results suggest a structure for full-length MOD1 where it dimerizes through the C–terminal domain alone, with the two N–terminal domains moving independently of each other at the end of flexible linkers.
The shadow chromo domain dimer binds to a single CAF MIR peptide
Both CAF1p150 (Murzina et al., 1999) and TIF1β (Le Douarin et al., 1996; Nielsen et al., 1999; Ryan et al., 1999) share a conserved peptide motif named MIR, for MOD1 interacting region, which is essential for their interaction with MOD1. To investigate the interaction between MOD1C and CAF1 further, we expressed a 66 amino acid peptide from mouse CAF1p150 (amino acids 204–269) as a His-tagged fusion protein in E.coli. This peptide comprises the overlapping sequence found in all of the fragments of CAF1 that were originally isolated in the two-hybrid screen with MOD1 (Murzina et al., 1999).
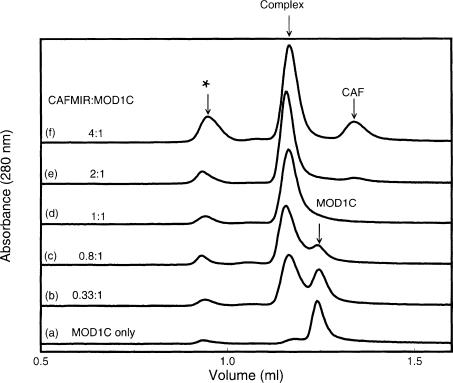
The stoichiometry of binding was studied using gel-filtration chromatography. Samples were analysed on a Superdex 75 column after mixing varying amounts of CAF MIR with a constant amount of MOD1C (Figure 5). A single peak, corresponding to the complex, was detected during gel filtration of a 1:1 mixture of CAF MIR to MOD1C dimer (Figure 5, trace d). Mixtures with lower CAF MIR:MOD1C dimer ratios showed an additional peak corresponding to free MOD1 (e.g. Figure 5, trace c), whilst mixtures with higher CAF MIR:MOD1C dimer ratios showed an additional peak corresponding to free CAF1 (e.g. trace e). At no ratio could we see all three peaks at the same time, indicating that the complex is stable during gel filtration. It is particularly noteworthy that, at a CAF MIR:MOD1C dimer ratio of 2:1 (trace e), we do see free CAF1. This would not be expected if two molecules of CAF1 bound to one molecule of MOD1C dimer, i.e. if one molecule of CAF1 bound to each subunit of the MOD1C dimer.
Fig. 5. Titration of MOD1C with the CAF1 MIR shows that one CAF1 peptide binds to one MOD1C dimer. The figure shows traces (_A_280) from gel filtration on a Superdex S75 column (2.4 ml) of MOD1C only (a) and different mixtures containing MOD1C dimer and CAF MIR in the ratios 1:0.33 (b), 1:0.8 (c), 1:1 (d), 1:2 (e) and 1:4 (f). The purity of the MOD1C and CAF MIR samples was checked by SDS–PAGE prior to mixing the proteins in the appropriate ratios. The arrows mark the positions at which free MOD1C, free CAF MIR and their complex eluted from the column. A small peak was observed eluting in the void volume of the column, suggesting that a small amount of high-molecular-weight aggregates was present in the mixtures. (Note that CAF MIR absorbs less strongly than MOD1C at 280 nm.)
The results of the gel filtration suggest that one molecule of CAF MIR binds to one MOD1C dimer. To confirm this we performed equilibrium sedimentation analysis of a 25 residue MIR-containing peptide from CAFp150 (amino acids 211–235, _M_r 2736 Da), MOD1C, and the complex of the two. The synthetic peptide was chosen for analytical centrifugation because the larger CAF MIR fragment (residues 204–269) was prone to both degradation and aggregation. MOD1C was mixed with an excess of CAF MIR 25mer and the mixture was eluted through an S75 gel-filtration column to remove excess peptide. Ultracentrifugation gave a molecular weight of 2.93 kDa for the peptide, 16.63 kDa for the MOD1C dimer and 19.17 kDa for the complex. This agrees with the gel-filtration results. [Note that a larger synthetic peptide (25mer) was used here to give a greater molecular weight difference than would have been obtained with the minimal 13mer peptide, see below.]
Definition of the MIR peptide binding site on the shadow chromo domain
We next sought to identify the MOD1C residues involved in the interaction with mouse CAF1p150. A CAF MIR 13mer (residues 220–232 of mouse CAF1p150), containing only the essential conserved peptide motif found in CAF1 and TIF1β (Murzina et al., 1999), was used in NMR experiments to map its binding site on MOD1C.
The 1H and 15N chemical shifts in 2D 1H–15N HSQC spectra of 15N-labelled MOD1C, before and after complex formation with excess unlabelled peptide, were determined to identify residues that are affected by ligand binding or conformational changes. As the majority of cross peaks in the spectrum of the complex were significantly perturbed, a 3D 15N-separated NOESY-HSQC spectrum was used to assign as many of the cross peaks as possible. Of the 58 non-proline residues, whose amide protons do not exchange rapidly with the solvent, 15 had cross peaks that did not change on the addition of peptide. A further 27 gave rise to two HSQC cross peaks of approximately equal intensity, both of which were shifted relative to their position in the isolated protein. For another three residues only one highly perturbed cross peak could be found (the other presumably being too shifted to be easily identified). Finally, the remaining 13 residues could not be identified, their chemical shifts also being too perturbed for analysis with data from this spectrum alone. An estimate of the number of HSQC peaks that remain unassigned suggests, however, that these residues are also likely to give rise to two HSQC cross peaks each.
The magnitude of the perturbations in the spectra that are observed upon binding is consistent with the formation of a tight complex and points to some changes in the structure of the protein on complex formation. The residues whose shifts are most strongly perturbed, and therefore most likely to be close to the ligand, form a contiguous region on the surface of the dimer. They lie in the C–terminal end of the second helix (helix α2), the C–terminal tail, the first β–strand and the first half of the second β–strand (see Figure 6). Many of the residues in the protein give rise to two different, perturbed, but identifiable HSQC cross peaks, suggesting that the same residue in the two different MOD1C subunits experiences a different local environment. This is not in itself unexpected, as a complex between a symmetric dimer and a single, non-symmetric peptide must of necessity be asymmetric. It is, however, surprising that the asymmetry is observed over so large a part of the molecule. Neither the NMR nor gel-filtration experiments point to the presence of more than one species in solution, and the presence of free MOD1C can be specifically excluded. There is no sign of free MOD1C in the NMR spectra and its presence is not compatible with a _K_d for complex formation of 2 μM, as determined by fluorescence spectroscopy (data not shown).
Fig. 6. Mapping of the MIR peptide binding site on MOD1C. The molecular surface of the MOD1C dimer is shown (A), together with a cartoon of the structure (B). (The view shown is related to that in Figure 3 by a 90° rotation about the vertical axis.) Residues for which there are no data are in white, unperturbed residues are in light blue, residues for which there are two cross peaks are in blue, residues for which only one highly perturbed cross peak could be found are in magenta and the most highly perturbed residues are in red. The position of the Trp170 side chain in the binding site is shown in yellow (see the text for details). The surface plot was made using GRASP (Nicholls et al., 1991).
TIF and CAF MIR compete for the MOD1C binding site
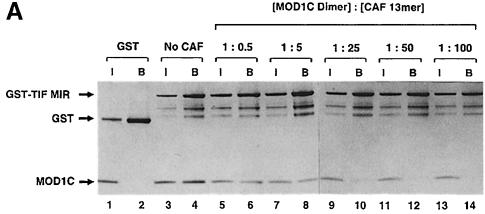
To investigate whether the TIF and CAF MIRs interact with the same binding site on the shadow chromo domain, we studied the binding of MOD1C to TIF1 MIR in the presence and absence of the CAF MIR 13mer peptide. TIF1 MIR [residues 449–567 of TIF1β expressed as a glutathione _S_–transferase (GST) fusion protein; Murzina et al., 1999] was bound to glutathione–agarose and used to pull down recombinant MOD1C (Figure 7A, lanes 3 and 4). The addition of increasing concentrations of CAF MIR 13mer progressively reduced, and in the end abolished, MOD1C binding to the GST–TIF MIR fusion protein (Figure 7A, lanes 5–14).
Fig. 7. (A) The MIR regions of TIF1β and CAF1 compete for binding to MOD1C. GST–TIF MIR was immobilized on glutathione–agarose beads and mixed with recombinant MOD1C. After extensive washing in buffer A150 (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 10% glycerol and 0.05% NP–40) the proteins remaining on the beads were separated using SDS–PAGE. The interaction between MOD1C and GST–TIF MIR was investigated in the presence of increasing amounts of a CAF1 13mer peptide containing the conserved MOD1C-binding motif. The input lanes (I) represent 20% of the amount of MOD1C present in the assay mixture, while the bound lanes (B) represent the total amount of MOD1C that remained bound to GST–TIF MIR following washing. The amounts of MOD1C and GST–TIF MIR were kept constant in each assay. (B and C) MOD1C mutants do not bind to the CAF1 MIR and TIF MIR peptides. Wild-type and mutant MOD1C proteins were in vitro translated using the TNT T7 Quick Coupled Transcription/Translation kit (Promega). Similar amounts of each 35S-labelled MOD1C protein were incubated with recombinant GST–CAF MIR (B) and GST–TIF MIR (C). After extensive washing, the proteins remaining on the beads were separated by SDS–PAGE, Coomassie Blue stained and detected by autoradiography. Auto- radiographs of the gel are presented in the figure. Lanes labelled I contain the equivalent of 20% of the input proteins. Lanes labelled B show proteins eluted from the glutathione–agarose beads following washing. Parts of the Coomassie-stained gels showing the amount of GST fusions or of GST (lanes 13 and 14) in the binding assays are presented in the bottom panels.
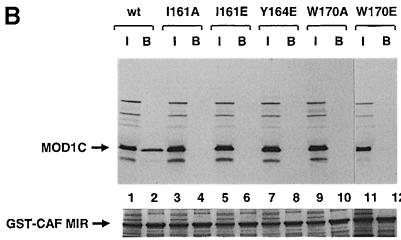
To investigate the competition further, we determined the effect of the W170A and W170E MOD1C mutations on peptide binding. W170 is one of the residues most affected by peptide binding in the NMR studies and is located at the centre of the mapped binding region (see Figure 6). Wild-type MOD1C and the W170A and W170E mutants were translated in vitro and assayed for their ability to bind to the GST–MIR peptides. The TIF and CAF MIR fragments obtained in the yeast two-hybrid screen for proteins interacting with MOD1 (Murzina et al., 1999) were expressed and purified as GST fusion proteins in E.coli for these experiments. As shown in Figure 7, neither the W170A nor the W170E mutant was able to bind to either the CAF MIR (Figure 7B, lanes 9–12) or the TIF MIR peptide (Figure 7C, lanes 9–12). However, the wild-type MOD1C did bind both (Figure 7B, lanes 1 and 2, and 7C, lanes 1 and 2), showing that W170 is involved in the binding of both MIRs. These experiments strongly suggest that the CAF and TIF MIR peptides bind to the same site on MOD1C.
A dimeric shadow chromo domain is required for the interaction with a MIR peptide
To investigate whether a dimer structure is important for the interaction we studied the binding of the monomeric I161A, I161E and Y164E MOD1C mutants to the CAF and TIF MIR peptides. I161 and Y164 are buried deep in the dimer interface and are unlikely to mediate the interaction directly with the CAF MIR peptide (Figure 3C). Moreover, the NMR spectrum of the complex indicates that the local environment of these residues does not change much upon peptide binding (coloured dark blue in Figure 6). In vitro translated wild-type MOD1C as well as the monomeric mutants were assayed for their ability to bind to the GST–MIR peptides. As shown in Figure 7, only the wild-type MOD1C bound to GST fusion proteins containing the MIRs from CAF1p150 (Figure 7B, lanes 1 and 2) or TIF1β (Figure 7C, lanes 1 and 2); none of the monomeric mutants bound (Figure 7B and C, lanes 3–8). In the control, wild-type MOD1C did not bind to GST alone, showing that the interaction with the MIR peptides was specific. To confirm that the failure of the mutants to bind was not due to the in vitro translated proteins being misfolded, we repeated the experiment using recombinant, E.coli expressed I161E and Y164E MOD1C whose monomeric and folded state had been demonstrated previously. The recombinant mutants did not bind to the CAF or TIF MIR GST fusion proteins (data not shown). These studies demonstrate that peptide binding depends on the formation of the MOD1C dimer.
Discussion
The overall structure of the MOD1 protein
Sequence alignment of the HP1 family proteins suggested that they consist of an N–terminal chromo domain and a C–terminal shadow chromo domain connected by a less conserved linker that is rich in charged residues. Our previous limited proteolysis data showed that both the linker and the N–terminus were very accessible to proteases, but that the two domains were resistant, the C–terminal being the more so (Ball et al., 1997). We expressed the defined domains in E.coli and determined the structure of both the chromo (Ball et al., 1997) and the shadow chromo domains (this work).
We find that the C–terminal shadow chromo domain of MOD1 forms a tight homodimer (Figure 3), and sedimentation analysis suggests that the upper limit for the dissociation constant is <150 nM (data not shown). The shadow chromo domain has the same fold as the interleukin–8 family of proteins, many of which form homodimers either by exchanging their helices or via interactions between their N–termini. The shadow chromo domain, however, exhibits a novel mode of dimerization in which the helices of one monomer interact with those of the other.
The structure of each subunit of MOD1C bears a striking resemblance to that of the N–terminal chromo domain that we determined previously (Ball et al., 1997) (see Figure 3B). Most amino acid residues forming the hydrophobic core of the MOD1C shadow domain are conserved not only between the different shadow domains, but also between the shadow and chromo domains (Figure 1). The structures can be superimposed over 25 residues in the β–sheet (N, 24–43 and 51–55; C, 120–139 and 145–149) with a root-mean-square deviation (r.m.s.d.) for the Cα atoms of 0.97 Å; those residues that are most highly conserved between the shadow and chromo domains occupy structurally similar positions.
We have used 15N relaxation experiments to study the dynamics of the protein's backbone amides to understand whether the individual domains interact with each other in the intact protein. No evidence of an interaction between the N–terminal chromo domain and either the C–terminal shadow domain or the other N–terminal chromo domain was found (Figure 2). Taken together with previous limited proteolysis data, the results show that the intact MOD1 protein is a dimer in which the N–terminal chromo domains are attached to the C–terminal, dimeric, shadow chromo domain by flexible linkers.
A plausible model for HP1 function might involve the N–terminal chromo domain being required for localization, such that the C–terminal shadow chromo domain can recruit other proteins to act at the appropriate location in chromatin. So far, however, yeast two-hybrid, in vitro experiments and phage display approaches have not identified functional partners of the chromo domain, apart from the ORC. It is possible that some post-translational modification, either in the chromo domain or in the unknown partner(s), is required. On the other hand, many different partners of the shadow chromo domain have been identified—the two interaction domains in HP1 proteins thus provide great flexibility for the localization of different functional complexes at distinct sites in the nucleus.
Interactions between HP1 proteins
In principle, the HP1 family proteins might interact with each other to form higher order complexes in two different ways. First, the HP1 dimers might interact with each other to form higher order complexes, e.g. tetramers, or alternatively, different HP1 monomers might interact to form heterodimers.
We could not detect any sign of further specific multimerization of MOD1, either by gel-filtration chromatography or sedimentation analysis. To test whether the HP1 proteins might form higher multimeric states, e.g. tetramers with either itself or other HP1 proteins, we attempted to pull down recombinant full-length MOD1 using recombinant GST fusions of HP1α, MOD1 and MOD2. In no case could we detect interactions (data not shown). The low dissociation constant that we observe for MOD1 suggests that all our recombinant proteins would be present as homodimers before we mixed them. Thus, these experiments are not complicated by the possibility of heterodimer formation (see below). However, one cannot rule out the possibility of further multimerization of the protein upon post-translational modification, such as phosphorylation, which is known to occur in eukaryotic cells (Minc et al., 1999; Zhao and Eissenberg, 1999).
The unmodified HP1 proteins could nevertheless form heterodimers directly with one another. Most residues involved in the dimer interface are conserved between the shadow domains of different HP1 proteins, e.g. A125, L132, N153, P157, I161, Y164, L168 and W170 are conserved in all except swi6. In addition, interactions have previously been reported between mouse HP1α and either itself or MOD1 in a yeast two-hybrid screen (Le Douarin et al., 1996). Moreover, human HP1α binds to both itself and to HP1γ in pull-down assays using in vitro translated proteins (Ye et al., 1997). Based on the biochemical, sequence and structural data, there is therefore a possibility of heterodimer formation between different HP1 mono– mers. Nevertheless, so far no biological functions have been ascribed to such interactions and it has been shown that HP1α, HP1β and HP1γ generally behave and localize differently in mammalian cells (Minc et al., 1999).
Shadow chromo domain interactions with other proteins
We have mapped the region of MOD1C involved in the interaction with proteins containing the MIR motif by NMR. The residues involved comprise the C–terminal end of the second helix, the C–terminal tail and the adjacent residues from the first and second β–strands (Figure 6). W170, which is located at the centre of this region, appears to play a critical role in the interaction. Its mutation to either A or E abolishes MOD1C binding to both the TIF and CAF MIRs (Figure 7B and C), but does not affect the dimeric nature of the domain (Figure 4).
The gel-filtration and sedimentation analyses demonstrate that one MIR peptide binds to one shadow chromo domain dimer (Figure 5). Given this stoichiometry, one would expect that interaction with the peptide would induce asymmetry in the dimer, and indeed we see strong evidence for this in the NMR spectra. We imagine two possible modes of peptide binding to MOD1C. In the first, one of the monomers binds a peptide molecule, and this binding prevents the other monomer from binding a second peptide, e.g. by allosteric changes. Alternatively, both monomers might be involved in binding a single peptide molecule. We favour the latter possibility because we found that the monomeric MOD1C mutants are not able to bind to the TIF and CAF MIR fragments (Figure 7B and C). We interpret the inability of these monomeric mutants to bind the CAF MIR peptide as being due to disruption of the binding surface, where the peptide binds to both subunits at the dimer interface.
This mode of protein–protein interaction in which a single monomeric peptide is recognized by a dimeric protein interaction motif is unprecedented in intracellular proteins. The only similar example occurs in the major histocompatibility complex (MHC II), where a single peptide binds to a site formed by two different polypeptide chains. The mapped region does not have any deep groove or cavity that would allow us to propose a detailed mode of binding. One possibility is that it binds in an extended conformation to the N–terminal β–strand in the shadow chromo domain (thereby extending the β–sheet) and at the same time makes contact with residues in the C–terminal tail of the other subunit. Another possibility is that the MIR peptide binds in between the C–terminal tails, moving them apart and thereby creating the necessary cavity for binding. This would be consistent with the NMR data which suggest that the C–terminal tail is not as well structured as the rest of the domain and such a mode of binding would explain why a large part of the molecule becomes asymmetric upon peptide binding. Clearly, the detailed mode of binding will need to await solution of the 3D structure of the complex.
The shadow chromo domain has recently been demonstrated to bind peptides related to the MIR consensus in phage display experiments (Smothers and Henikoff, 2000). However, the authors' suggestion that the peptides mimic the PQVVI sequence found in the C–terminal helix, and thereby disrupt the dimer, is not borne out by our work. We observe only slight chemical shift perturbations in this region of the protein on peptide binding, consistent with the fact that only the I is involved in significant inter-monomer interactions, whilst the two Vs are buried within the monomer.
The TIF and CAF MIRs compete for binding to MOD1C
Given their sequence similarity, we thought it possible that the TIF1β and CAF1p150 MIR peptides would interact with the same binding site on MOD1C and the experiments shown in Figure 7 support this view. Whilst the conserved MIR motif is capable of binding to MOD1C, additional adjacent residues are also involved (data not shown). Given the lack of sequence similarity between TIF1β and CAF1p150 outside the conserved MIR motif, we speculate that the flanking regions of the two proteins might bind differently to the shadow chromo domain. Structural studies of the two complexes will reveal which MOD1 residues are involved and this might enable the design of MOD1 mutants that would bind specifically to either CAF1p150 or TIF1β, allowing further insight into the biological roles of each complex.
Some of the proteins that interact with MOD1C do not possess a recognizable MIR motif and may bind in a different way, or even to a different binding site. A possible location for an alternative site might be at the other end of the dimer axis where there is a relatively hydrophobic surface patch, made up of residues V154, P157 and Q158.
Regulation of HP1 protein interactions
HP1 proteins are phosphorylated in vivo and this phosphorylation may be an important mechanism for regulating the proteins' multimerization and/or interactions. Zhao and Eissenberg (1999) have identified three casein kinase II phosphorylation sites on Drosophila HP1, S15, S199 and S202, which are needed for heterochromatin binding. In HP1β and HP1γ, but not HP1α, T169 and S172 lie in a similar sequence context to that of the C–terminal phosphorylation sites in Drosophila HP1. T169 is located close to the MIR binding site on the shadow chromo domain, suggesting that its phosphorylation might prevent binding by the hydrophobic MIR peptides. Our studies of the structure and interactions of MOD1C thus suggest a mechanism by which phosphorylation might alter the function of HP1 proteins during the cell cycle and/or development, where it is known that their phosphorylation patterns change.
Materials and methods
DNA manipulations
The constructs and strains for E.coli expression of His-tagged MOD1 (residues 1–185) and MOD1C (residues 104–171) have been described previously (Ball et al., 1997; Murzina et al., 1999). Site-directed mutagenesis of wild-type mouse MOD1C (residues 104–171) in a pET16b construct (Novagen) was performed using QuickChange (Stratagene) according to the manufacturer's instructions. For expression of GST–CAF MIR (residues 176–327) and GST–TIF MIR (residues 449–567), cDNA fragments obtained from a two-hybrid screen (Murzina et al., 1999) were subcloned directly into the _Bam_HI and _Eco_RI sites of the pGEX-5X vector (Pharmacia). The region of CAF1p150 cDNA common to all the p150 clones identified in the two-hybrid screen (Murzina et al., 1999) was subcloned by PCR and ligated into the _Nde_I and _Bam_HI sites of the pET16b vector (Novagen).
Expression and purification of MOD1 and MOD1C
Unlabelled, 15N- and 15N/13C-labelled proteins were expressed and purified as described (Ball et al., 1997), with the following additional steps. After Factor Xa cleavage of the His-tag, the MOD1 or MOD1C samples were separated from the His-tag and further purified by MonoQ HR 5/5 ion-exchange and Superdex S75 gel-filtration chromatography (Pharmacia), using standard protocols. (Note that the final purified proteins have an additional N–terminal histidine and methionine residue, which originate from the vector.)
A mixed dimer sample for NMR spectroscopy was made by mixing equal amounts of unlabelled and 15N/13C-labelled MOD1C in a large volume of 6 M guanidinium hydrogen chloride (GuHCl), 10 mM sodium phosphate pH 8.0, 1 mM dithiothreitol (DTT) and 1 mM EDTA. The protein sample was renatured by serial dilution with the same buffer containing decreasing amounts of GuHCl (4.5, 4, 3 and 0 M GuHCl) prior to dialysis into NMR buffer.
Expression and purification of CAF MIR and TIF MIR proteins
For the pull-down assays, GST–CAF and GST–TIF MIRs were expressed and purified using standard protocols. For the binding studies of CAF MIR to MOD1C by gel-filtration chromatography, His-tagged CAF MIR (residues 204–269) was expressed in E.coli JM109 (DE3) cells. The peptide was affinity purified under denaturing conditions (in the presence of 6 M GuHCl or 8 M urea) using a Ni–NTA column (Qiagen), and further purified by gel filtration under native conditions using a Superdex 75 column (Pharmacia).
NMR spectroscopy and spectral assignments
NMR spectra were recorded on Bruker 500, 600 and 800 MHz spectrometers at either 30 or 35°C. Protein samples contained between 0.6 and 1.4 mM MOD1C monomer in 10 mM sodium phosphate buffer at pH 8.0, containing 10 mM perdeuterated DTT, 0.05% sodium azide and either 10 or 100% D2O. Resonance assignments were achieved using standard triple resonance and homonuclear methods. NOE data for the structure calculations were mainly obtained from 2D homonuclear NOESY spectra in H2O and D2O, as well as 3D 15N- and 3D 13C-separated NOESY-HSQC spectra in H2O, all recorded with mixing times of 100–120 ms. In addition, a 3D 15N-separated NOESY-HSQC spectrum recorded with a 40 ms mixing time was employed. 15N relaxation measurements were performed at 600 MHz for the N- and C–terminal domains and at 500 MHz for full-length MOD1. All NMR data were processed with the program AZARA (W.Boucher, unpublished data) and analysed using ANSIG (Kraulis, 1989; Kraulis et al., 1994).
Structure calculations
Distance restraints were derived from 4182 cross peaks, of which 2334 were assigned manually and 1848 were assigned based on their chemical shifts using ‘Connect’ from AZARA. Cross peaks were grouped (according to intensity) as strong, medium, weak and very weak, the corresponding restraint limits being 0.0–2.7, 0.0–3.3, 0.0–5.0 and 0.0–6.0 Å, respectively. A 3D 13C/15N-filtered, 13C-separated NOESY-HSQC (Zwahlen et al., 1997) and a 2D 13C/15N-double-half-filtered NOESY experiment (Folmer et al., 1995) (mixing times 150 ms) gave rise to cross peaks that were known to be either inter- or intra-monomer. After removal of mutually redundant restraints, there remained a total of 58 cross peaks known to be inter-monomer, 426 cross peaks known to be intra-monomer and 1606 cross peaks that were ambiguous, i.e. either inter- or intra-monomer. In addition, 21 ambiguous distance restraints were incorporated to constrain hydrogen bonds (identified as exchanging slowly with water) to be within 2.5 Å of a hydrogen bond acceptor.
The structures were iteratively refined in CNS 0.9 (Brunger et al., 1998) using the PARALLHDG v5.1 forcefield in PROLSQ mode (Linge and Nilges, 1999) and ARIA (Nilges et al., 1997). To deal with the dimeric nature of the protein, axially symmetric starting structures were generated from random coordinates for one monomer and subsequently rotated by 180° around an axis 5 Å from their centre of mass to generate the other monomer. To maintain axial symmetry throughout the calculations, the non-crystallographic symmetry restraint was applied with weight 10, and distance-based symmetry restraints were used (O'Donoghue et al., 1996).
The quality of the calculated structures was assessed with PROMOTIF (Hutchinson and Thornton, 1996) and PROCHECK (Laskowski et al., 1993).
PDB code
The coordinates have been deposited in the Brookhaven Protein Data Bank under accession number 1dz1.
Acknowledgments
Acknowledgements
We thank Michael Nilges (EMBL, Heidelberg) for CNS and X-PLOR scripts, Jo Butler (MRC LMB, Cambridge) and Matthew Deacon for the MOD1 sedimentation analysis, Len Packman and the PNAC Facility for peptide synthesis, amino acid analysis, mass spectrometry and oligonucleotide synthesis, and Alexey Murzin for helpful discussions. We thank the Wellcome Trust for financial support and the analytical ultra-centrifuge. The Cambridge Centre for Molecular Recognition and the National 800 MHz NMR Facility are supported by the BBSRC and the Wellcome Trust.
References
- Aagaard L. et al. (1999)Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J., 18, 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aasland R. and Stewart, A.F. (1995) The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res., 23, 3168–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball L.J. et al. (1997)Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J., 16, 2473–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst R.W., Hardman, C.H., Thomas, J.O. and Laue, E.D. (1995) Backbone dynamics of the A-domain of HMG1 as studied by 15N NMR spectroscopy. Biochemistry, 34, 16608–16617. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998)Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cavalli G. and Paro, R. (1998) Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol., 10, 354–360. [DOI] [PubMed] [Google Scholar]
- Cleard F., Delattre, M. and Spierer, P. (1997) SU(VAR)3-7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. EMBO J., 16, 5280–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J.C., James, T.C., Foster-Hartnett, D.M., Hartnett, T., Ngan, V. and Elgin, S.C. (1990) Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 87, 9923–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J.C., Ge, Y.W. and Hartnett, T. (1994) Increased phosphorylation of HP1, a heterochromatin-associated protein of Drosophila, is correlated with heterochromatin assembly. J. Biol. Chem., 269, 21315–21321. [PubMed] [Google Scholar]
- Ekwall K., Javerzat, J.P., Lorentz, A., Schmidt, H., Cranston, G. and Allshire, R. (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science, 269, 1429–1431. [DOI] [PubMed] [Google Scholar]
- Fanti L., Giovinazzo, G., Berloco, M. and Pimpinelli, S. (1998) The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell, 2, 527–538. [DOI] [PubMed] [Google Scholar]
- Folmer R.H.A., Hilbers, C.W., Konings, R.N.H. and Hallenga, K. (1995) A C-13 double-filtered NOESY with strongly reduced artifacts and improved sensitivity. J. Biomol. NMR, 5, 427–432. [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Fredericks, W.J., Jensen, D.E., Speicher, D.W., Huang, X.P., Neilson, E.G., Rauscher, F.J., III (1996) KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev., 10, 2067–2078. [DOI] [PubMed] [Google Scholar]
- Hutchinson E.G. and Thornton, J.M. (1996) PROMOTIF—a program to identify and analyze structural motifs in proteins. Protein Sci., 5, 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T.C. and Elgin, S.C. (1986) Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol., 6, 3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R. and Alberts, B.M. (1995) Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J. Cell Sci., 108, 1419–1431. [DOI] [PubMed] [Google Scholar]
- Kellum R., Raff, J.W. and Alberts, B.M. (1995) Heterochromatin protein 1 distribution during development and during the cell cycle in Drosophila embryos. J. Cell Sci., 108, 1407–1418. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Zhou, S. and Lucchesi, J.C. (1995) The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res., 23, 4229–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1989) ANSIG: a program for the assignment of protein 1H 2D NMR spectra by interactive graphics. J. Magn. Reson., 24, 627–633. [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Kraulis P.J., Domaille, P.J., Campbell-Burk, S.L., Van Aken, T. and Laue, E.D. (1994) Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry, 33, 3515–3531. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur, M.W., Moss, D.S. and Thornton, J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Le Douarin B. et al. (1995)The N–terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J., 14, 2020–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B., Nielsen, A.L., Garnier, J.M., Ichinose, H., Jeanmougin, F., Losson, R. and Chambon, P. (1996) A possible involvement of TIF1 α and TIF1 β in the epigenetic control of transcription by nuclear receptors. EMBO J., 15, 6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Linge J.P. and Nilges, M. (1999) Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J. Biomol. NMR, 13, 51–54. [DOI] [PubMed] [Google Scholar]
- Lorentz A., Ostermann, K., Fleck, O. and Schmidt, H. (1994) Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene, 243, 139–143. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon, D.J. (1997) Raster3D photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Minc E., Allory, V., Worman, H.J., Courvalin, J.C. and Buendia, B. (1999) Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma, 108, 220–234. [DOI] [PubMed] [Google Scholar]
- Moosmann P., Georgiev, O., Le Douarin, B., Bourquin, J.-P. and Schaffner, W. (1996) Transcriptional repression by RING finger protein TIF1β that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res., 24, 4859–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina N., Verreault, A., Laue, E. and Stillman, B. (1999) Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell, 4, 1–20. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp, K.A. and Honig, B. (1991) Protein folding and association—insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nielsen A.L., Ortiz, J.A., You, J., Oulad-Abdelghani, M., Khechumian, R., Gansmuller, A., Chambon, P. and Losson, R. (1999) Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J., 18, 6385–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilges M., Macias, M.J., O'Donoghue, S.I. and Oschkinat, H. (1997) Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from β-spectrin. J. Mol. Biol., 269, 408–422. [DOI] [PubMed] [Google Scholar]
- O'Donoghue S., King, G.F. and Nilges, M. (1996) Calculation of symmetric multimer structures from NMR data using a priori knowledge of the monomer structure, co-monomer restraints and interface mapping: the case of leucine zippers. J. Biomol. NMR, 8, 193–206. [DOI] [PubMed] [Google Scholar]
- Pak D.T., Pflumm, M., Chesnokov, I., Huang, D.W., Kellum, R., Marr, J., Romanowski, P. and Botchan, M.R. (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell, 91, 311–323. [DOI] [PubMed] [Google Scholar]
- Paro R. (1990) Imprinting a determined state into the chromatin of Drosophila. Trends Genet., 6, 416–421. [DOI] [PubMed] [Google Scholar]
- Paro R. and Hogness, D.S. (1991) The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl Acad. Sci. USA, 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero J.S., Hartnett, T. and Eissenberg, J.C. (1995) Functional analysis of the chromo domain of HP1. EMBO J., 14, 3977–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remboutsika E., Lutz, Y., Gansmuller, A., Vonesch, J.L., Losson, R. and Chambon, P. (1999) The putative nuclear receptor mediator TIF1 α is tightly associated with euchromatin. J. Cell Sci., 112, 1671–1683. [DOI] [PubMed] [Google Scholar]
- Ryan R.F., Schultz, D.C., Ayyanathan, K., Singh, P.B., Friedman, J.R., Fredericks, W.J., Rauscher, F.J., III (1999) KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol., 19, 4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W.S. et al. (1993)Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. J. Cell Sci., 104, 573–582. [DOI] [PubMed] [Google Scholar]
- Singh P.B., Miller, J.R., Pearce, J., Kothary, R., Burton, R.D., Paro, R., James, T.C. and Gaunt, S.J. (1991) A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res., 19, 789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers J.F. and Henikoff, S. (2000) The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol., 10, 27–30. [DOI] [PubMed] [Google Scholar]
- Ye Q., Callebaut, I., Pezhman, A., Courvalin, J.C. and Worman, H.J. (1997) Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem., 272, 14983–14989. [DOI] [PubMed] [Google Scholar]
- Zhao T. and Eissenberg, J.C. (1999) Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J. Biol. Chem., 274, 15095–15100. [DOI] [PubMed] [Google Scholar]
- Zwahlen C., Legault, P., Vincent, S.J.F., Greenblatt, J., Konrat, R. and Kay, L.E. (1997) Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage λ N-peptide/boxB RNA complex. J. Am. Chem. Soc., 119, 6711–6721. [Google Scholar]