Analysis of the Human Endogenous Coregulator Complexome (original) (raw)
. Author manuscript; available in PMC: 2012 May 27.
Summary
Elucidation of endogenous cellular protein-protein interactions and their networks is most desirable for biological studies. Here we report our study of endogenous human coregulator protein complex networks obtained from integrative mass spectrometry-based analysis of 3,290 affinity purifications. By preserving weak protein interactions during complex isolation and utilizing high levels of reciprocity in the large dataset we identified many unreported protein associations, such as a transcriptional network formed by ZMYND8, ZNF687 and ZNF592. Furthermore, our work revealed a tiered interplay within networks that share common proteins, providing a conceptual organization of a cellular proteome composed of minimal endogenous modules (MEMOs), functional uniCOREs and regulatory complex-complex interaction networks (CCIs). This resource will effectively fill a void in linking correlative genomic studies with an understanding of transcriptional regulatory protein functions within the proteome for formulation and testing of new hypotheses.
Introduction
Protein-protein interactions constitute the molecular backbone of cell biology, where select proteins assemble into meta-stable complexes to form bioactive units (Alberts, 1998; Kocher and Superti-Furga, 2007). These complexes then dynamically associate with each other in context of larger networks to carry out diverse biological functions. Thus, understanding the basic mechanisms of cell homeostasis requires both knowledge of the composition of protein complexes and the interactions between them.
A systems biology view of protein interactions has begun to emerge from large-scale studies in model organisms such as yeast, worms, and fruit flies (Gavin et al., 2006; Giot et al., 2003; Ito et al., 2001; Krogan et al., 2006; Li et al., 2004; Uetz et al., 2000). These analyses were made possible due to the development of high throughput (HT) methods for measuring protein-protein interactions by affinity purification of tagged protein baits followed by mass spectrometry (AP/MS) and yeast-two-hybrid assays. Limitations in genetic manipulations hinder such studies in human cells. We thus developed a protocol for HT isolation and identification of endogenous protein complexes from human cell lines using primary antibody immunoprecipitation and mass spectrometry (IP/MS). We also addressed key limitations associated with such studies, which are cross-reactivity of primary antibodies and non-specific binding, and proposed an approach for the deconvolution of HT-IP/MS data into discrete protein complexes (Malovannaya et al., 2010).
Our current work was inspired by - and for a large part included - the Nuclear Receptor Signaling Atlas (NURSA) consortium proteomics effort, whose goal is to systematically isolate and identify the human nuclear receptor (NR) coregulator complexome. NR coregulators are a diverse group of molecules that associate with sequence-specific transcription factors to collectively modulate target gene expression (Lonard and O'Malley B, 2007; McKenna et al., 1999; O'Malley et al., 2008; Weake and Workman, 2010). Initial biochemical isolations of the mammalian coactivators Mediator and BAF/P-BAF, and HDAC corepressors revealed that many coregulators assemble into multi-subunit protein complexes (Gu et al., 1999; Guenther et al., 2000; Wang et al., 1996; Xue et al., 1998), implying that a comprehensive picture of the protein interaction networks is needed to better understand the regulation of biological processes in the cell.
This study presents the most extensive interaction dataset for endogenous regulatory human proteins obtained to date. By preserving both stable and weak protein interactions during complex isolations, we unveiled a modular and hierarchical organization of protein complex networks that serve as a blueprint for a better understanding of mechanisms of mammalian cell regulation. Our approach can be used in other biological systems as well; with broad applicability in mind we therefore discuss the analysis schema we used for the definition and annotation of protein complexes. Knowing the composition of protein complexes and their interaction networks ultimately will allow for an effective translation of genomic data to functional molecular biology. Such resources, therefore, have the potential to transform the field of basic research and greatly impact translational efforts.
Results and Discussion
A Comprehensive HT-IP/MS of the Human Endogenous Complexome
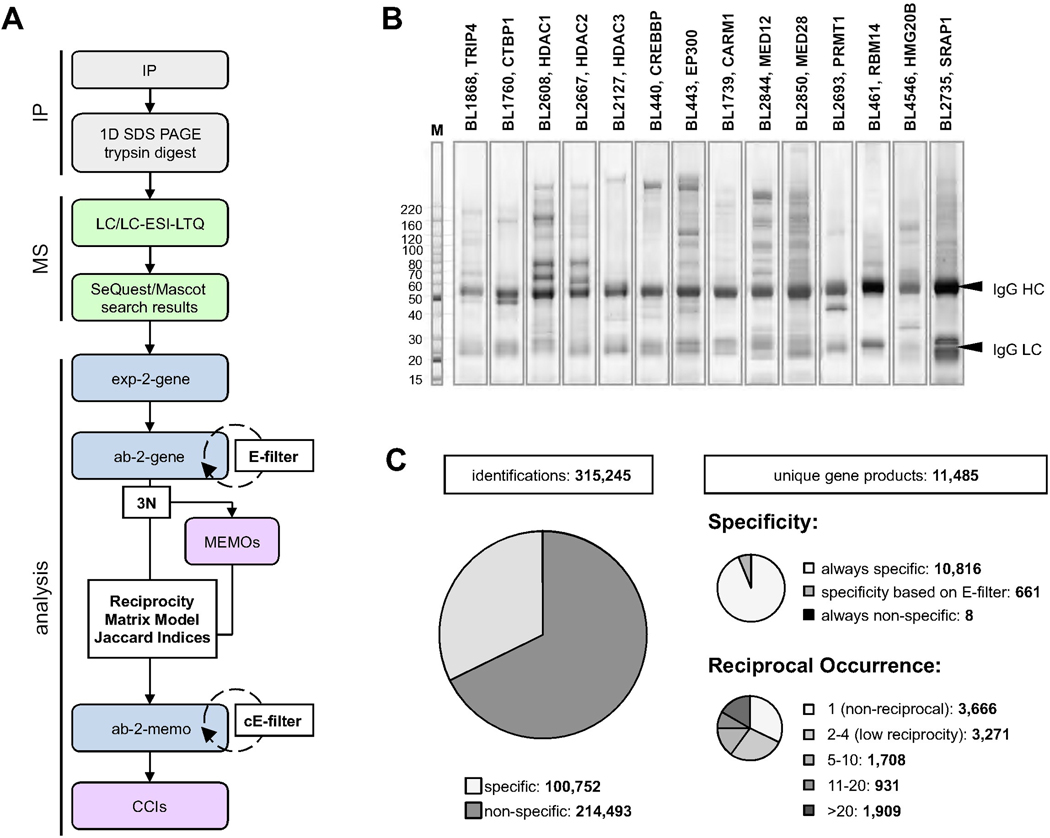
To initiate delineation of the ‘regulatory’ human protein complexome we preferentially targeted transcriptional and signaling proteins. We used 1,796 primary antibodies in 3,290 immunoprecipitation experiments (Figure 1A,B and Table S1, Table S2) to obtain a total of ~300,000 protein identifications, about 100,000 of which we deemed specific after in silico data filtering (Figure 1A,C). These identifications represent 11,485 unique human gene products, more than half of which we recovered redundantly using different antibodies.
Figure 1. HT-IP/MS Analysis of the Human Endogenous Complexome.
A) Our HT-IP/MS workflow consists of IP/MS followed by filtering of non-specific identifications, definition of minimal core complex modules (MEMOs), and assignment of complex-complex interactions (CCIs). Data relationships are abbreviated as ‘exp-2-gene’, ‘ab-2-gene’, and ‘ab-2-memo’ for experiment-to-genes, antibody-to-genes, and antibody-2-MEMOs, respectively. B) Representative SDS-PAGE of NR coregulator complexes. IgG HC and LC are heavy and light chains of primary antibodies. C) Approximately 40% of the human gene products were recovered in our HT-IP/MS data in ~100,000 specific protein identifications. Majority of these were found in at least two or more antibody-different experiments, laying a foundation for finding reciprocally verified protein associations.
To assess the overall discovery potential, we compared all possible unique pairs of protein associations from our dataset with human protein interactions found in the CORUM, BioGRID, IntAct, and HPRD resources (Figure S1). Our dataset covers about 70% of the protein associations reported in CORUM (Ruepp et al., 2010), while most of our high-confidence interactions are not listed in this nor any other current public repositories. Based on this comparison alone, our single-source study recovered the majority of protein complexes previously reported in the literature, but at the same time, significantly expanded the human interactome.
Deconvolution of the IP/MS Data Into Organized Cellular Complexome
The nature of IP/MS data output – a list of protein identifications without indications of specific interactions within and between complexes – commands a different logic for data analysis as compared to conventional protein-protein interaction (PPI) maps. We thus developed analysis schema that reveals an intrinsic tiered organization of the interactome in three discrete layers. These are (1) the minimal endogenous core complex modules (‘MEMOs’), (2) the unique core complex isoforms (‘uniCOREs’), and (3) the complex-complex interaction networks (CCIs) (Figure S2A). Such classification respects protein complex modularity and heterogeneity, which constitute a fundamental program of cellular organization, and aids in delineation of higher order protein interactions on a proteome-wide level.
Minimal Endogenous Modules (MEMOs) and Core Complex Isoforms (uniCOREs)
A core protein complex is loosely defined in biochemistry as strongly associated proteins that resist separation in column fractionations. Many core complexes, however, exhibit some level of variability in only a few components of otherwise similar protein assemblies. Such complex ‘isoforms’ can be isolated and shown to function exclusively of each other. For example, we recovered the MSL and NSL complexes that contain transcriptional regulator MYST1 histone acetyltransferase and recently have been shown to display different substrate specificities (Cai et al., 2010; Mendjan et al., 2006). To better convey such modularity, we identified variable components in protein complexes and formed a dataset of the minimal endogenous core complex modules (MEMOs). These modules represent invariant minimal complexes of proteins with stoichiometric interdependence across the entire IP/MS dataset and serve as the conceptual building blocks of the protein complexome. Ultimately, each of the human protein-coding gene products will be assigned to one MEMO only. MEMOs are then used to reconstitute all distinct protein complex ‘isoforms’, which we call unique cores (uniCOREs). The multi-subunit protein complexes conventionally described in the literature most closely correspond to uniCOREs in our resource and likely impart biological functional classification.
We recently have shown that high confidence protein interaction information can be derived from systematic IP/MS studies by enforcing two key constrains: reciprocal co-appearance of proteins in multiple antibody-different IPs and preservation of stoichiometry of the core components (Malovannaya et al., 2010). Operationally, MEMOs are identified by running reciprocal Near Neighbor Network (3N) analyses iteratively over the entire dataset to find sets of proteins with the highest correlations in different experiments. Proteins that do not show consistency in stoichiometric interactions form ‘singletons’, which are MEMOs with special implications discussed later.
De Novo Derivation of Complex-Complex Interactions
While much insight is gained from the core protein complexes, a more daunting task is to acquire information of the complex-complex interactions (CCIs). In fact, CCIs may represent the backbone of regulatory biology; yet, on no account have they been systematically addressed. We show here that CCIs can be derived de novo from HT-IP/MS data.
Transient interactions between protein complexes don’t show a stoichiometric dependence; we therefore used Boolean metrics on co-occurrences of MEMOs to find complex-complex interactions. Briefly, we first calculated reciprocity and a series of Jaccard indices for all protein associations (see Supplement). Furthermore, we adopted a matrix model score previously described as part of socio-affinity index (Gavin et al., 2006) to capture above-chance associations, and defined a CCI Rank that combines protein interaction scores for all subunits of a given MEMO as a chance indicator for true associations between MEMOs (Supplement). We have pre-calculated all CCIs for each gene product in our HT-IP/MS dataset and formed a resource of human endogenous MEMOs and CCI networks that allows researchers to interrogate our data. Subsequent iterative searching for different MEMO components can be used to identify uniCOREs, as described next.
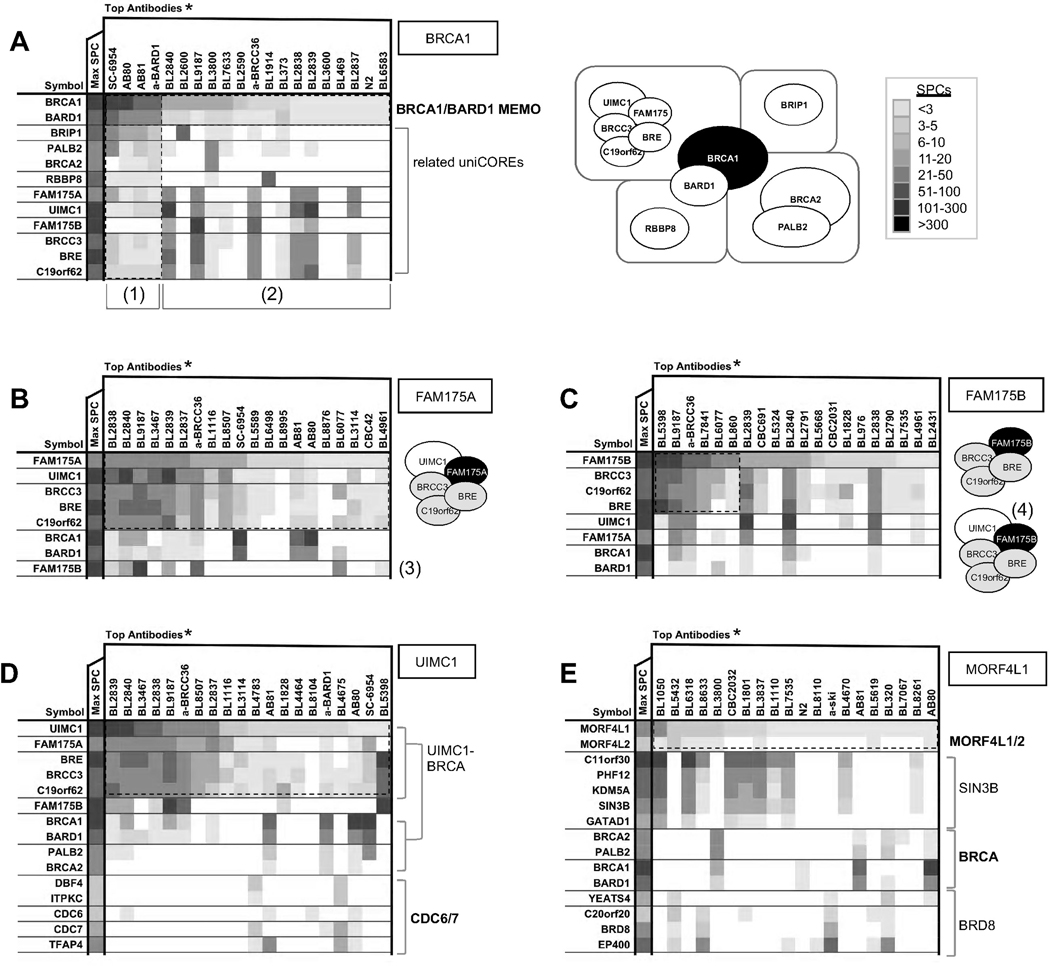
Example of Assignment of uniCOREs Through Reciprocal Exclusion in CCI Networks: The BRCA1 Extended Interaction Network
BRCA1 (breast cancer 1) has been extensively studied in DNA damage responses as well as transcriptional regulation. Multiple IP/MS experiments showed that the BRCA1 interactome is more heterogeneous and modular than previously thought. BRCA1 and BARD1 (BRCA1 associated RING domain 1) show stoicheometric interdependency and constitute a MEMO, and are found in a pool of apparently stable complexes either with BRCA1-interacting protein 1 (BRIP1), PALB2/BRCA2, RBBP8, BRE/BRCC3/C19orf62, FAM175A (Abraxas), FAM175B (Abraxas brother 1), or UIMC1 (Figure 2A and S2). These proteins do not have consistent stoichiometric relationships with each other, suggesting that they likely form different BRCA1 uniCOREs.
Figure 2. Protein Complex Heterogeneity in BRCA1 Network.
Partial BRCA1-related CCI networks, where individual MEMOs are separated by horizontal black lines, are shown to highlight the relationships between major components of the BRCA1 interactome (see also Figure S2). (*) Column headers specify antibody names, not the intended antigens that were used to generate the antibody. Despite conventions, it is often misleading to label IP/MS experiments with 'intended' antigens, because majority of antibodies cross-react with 2 or more proteins and some do not IP the intended antigen at all (see Supplemental Table S1). A) Discernible ‘hierarchical’ organization of protein interactions illustrated by a selection of IPs containing BRCA1 uniCOREs with the highest SPC identifications for all precipitated proteins (compare TopAntibodies lanes with MaxSPC lane). Although all proteins are equally true interactors of BRCA1, extensive reciprocal evidence visually implicates exclusive patterns: First three lanes are BRCA1 and BARD1 IPs (1), where all BRCA1-containing complexes are shared, while experiments that target specific uniCOREs (2) show non-uniform distributions of BRCA1 interactors. To the right is a schematic interpretation of the findings. B) Top experiments for FAM175A reveal a stoichiometric complex between FAM175A, UIMC1, and BRE/BRCC36/C19orf62, but not with FAM175B (3) C) 3N analyses for FAM175B (C) and UIMC1 (D) show that all IPs that have FAM175A predominantly contain UIMC1 and BRCA1 (4), and thus reveal autonomous FAM175A/B uniCOREs. E) MORF4L1/2 MEMO interacts with BRCA1/BARD1/PALB2/BRCA2 uniCORE, and also forms uniCOREs with chromatin remodeling complexes SIN3B and BRD8.
The interactions between UIMC1, FAM175A, FAM175B, and BRCC3/BRE/C19orf62 (‘BRCA1-A’ complex) have been described in the literature (Feng et al., 2009; Kim et al., 2007; Shao et al., 2009b; Wang et al., 2009a; Wang et al., 2007; Yan et al., 2007). In our dataset, FAM175A and FAM175B interact with UIMC1 in a mutually exclusive manner, and they resolve independently from each other in respective 3N analyses (Figure 2B,C). The FAM175A–UIMC1-BRCC3/BRE/C19orf62 uniCORE does not strongly bind known DNA damage response components, but co-precipitates CDC6/7 kinases, bridging into checkpoint BRCA1 functions (Figure 2D). In addition, we observed that this uniCORE preferentially interacts with the BRCA1/BARD1 MEMO, while the FAM175B–containing uniCORE can be found without UIMC1 and BRCA1 (Figure 2C). Specific functions of the latter complex, particularly in the BRCA1-independent context, are unknown.
Beyond UIMC1, BRCA1/BARD1 together with PALB2/BRCA2 bridge into mismatch repair processes via BRIP1 (‘BRCA1-B’ complex) and interactions with MLH1/PMS2 (Figure S2). BRCA1/BARD1/PALB2/BRCA2 also exists in a close CCI network with RBBP8 (‘BRCA1-C’ complex), the double-stranded break sensor MRN (MRE11-RAD50-NBS1) complex, CHEK1, RAD51, KEAP1 and MORF4L1/L2, implementing a BRCA DNA repair function. KEAP1, an E3 ubiquitin ligase adaptor (Cullinan et al., 2004; Kobayashi et al., 2004; Zhang et al., 2004), appears to be a new component of the BRCA1 interactome; its function in this network remains to be investigated. Interestingly, the MORF4L1/2 MEMO itself has at least two additional uniCOREs with transcriptional coregulator complexes SIN3B and BRD8 (Figure 2E), highlighting widespread heterogeneity in protein complex organization.
The BRCA example illustrates that interactome modules are separable by data analysis in silico. Moreover, a hierarchical representation of CCIs via MEMOs and uniCOREs is an appropriate way to illustrate these distinct, biologically relevant entities. As we continue to appreciate the modularity in protein complexes, it becomes increasingly sensible to catalogue the divergence of cellular networks anchored in proteins shared by multiple uniCOREs. In retrospect, the BASC complex we reported 10 years ago is actually a merged representation of BRCA1 CCI networks (Wang et al., 2000) that now also include other components (Feng et al., 2009; Kim and Chen, 2008; Shao et al., 2009a; Shao et al., 2009b; Zhang et al., 2009). Thus, our HT-IP/MS study allows a ‘systems view’ of the BRCA1 network, predicting various PPI dynamics and their vitality in biology. This is a testimony for the merit of a single-source HT-IP/MS effort, where uniform experimental conditions are applied and more consistent data are obtained for the analysis of context-dependent CCI networks.
Iterative Interrogation Reveals CCI Topology
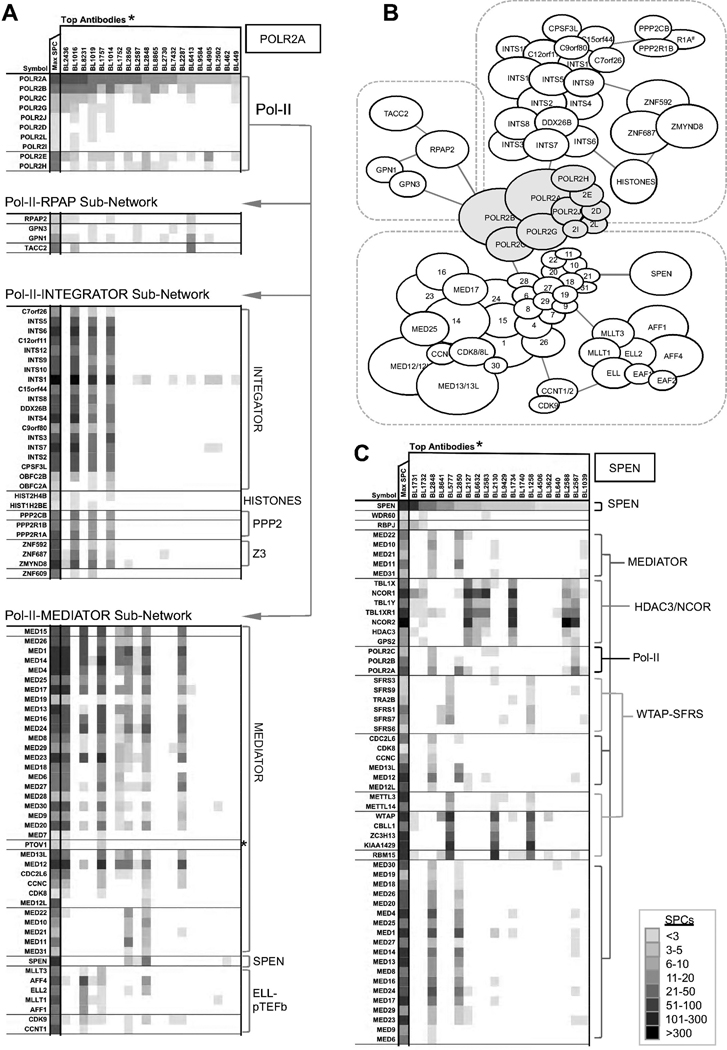
The most effective way to mine our data is to iteratively search for proteins within a particular CCI network to reveal nuances in network organization. For example, when the largest RNA polymerase Pol-II subunit (POLR2A) is used as the seed, six known Pol-II subunits are found along with most subunits of the Mediator and Integrator complexes, RPAP2, GPN1/GPN3, SPEN, ZMYND8/ZNF687/ZNF592, and CDK9/CCNT1 and ELL elongation complexes (Figure 3A,B). Reciprocal searches using MED15, INTS7, or RPAP2 all preferentially return Pol-II subunits, confirming these extensive interactions (Figure S3). Closer inspection of the data shows that the RPAP2 network lacks Mediator coactivator, but has GPN1/GPN3 and Integrator. Therefore, RPAP2 likely resides in a sub-network of Pol-II where Integrator and Mediator separate. Analysis of the MED15 data shows that this protein is found in two exclusive complexes: it is a part of the large Mediator complex, but also interacts separately with an E3-ligase TRIM11, which binds and degrades MED15 (Ishikawa et al., 2006). Our data imply that TRIM11 sequesters MED15 subunit away from the main body of the Mediator complex in a relatively stable and discrete complex. Accordingly, we have singled out MED15 as a separate MEMO to account for this exclusive interaction pattern.
Figure 3. Iterative Mining of HT-IP/MS Resource Reveals Topology of RNA Polymerase II Network.
A–B) 3N analyses for POLR2A show three separate subnetworks containing RPAP2-GPN1/3, Integrator complex, and Mediator complex (see also Figures S3 and S6). The MEMOs in the POLR2A network were grouped by patterns of their distribution to show that Mediator and Integrator sub-networks are vividly independent of each other. C) Coregulator SPEN stands on crossroads of three seminal transcriptional processes. Its CCI network suggests an association with Pol-II through Mediator or HDAC3/NCOR complex. SPEN also co-precipitates with proteins that regulate splicing process, including multi-subunit SFRS and WTAP complexes. (*) Heatmap column headers specify antibody names, not the intended antigens that were used to generate the antibody (for antibody data, see Supplemental Table S1).
Next we use SPEN (split ends homologue; a.k.a. SHARP) as an example of a single-coregulator MEMO at the crossroads of different transcriptional processes. SPEN CCI analysis reveals previously unappreciated protein interactions (Figure 3C). SPEN’s associations with Mediator explain its presence in the Pol-II network. This analysis also reveals the HDAC3/NCOR corepressor complex as a prominent interaction partner of SPEN. Finally, we recover SPEN interactions with a WTAP (Wilm’s tumor 1 associated protein) mRNA splicing complex, independently from HDAC3/NCOR and Mediator. Collectively, SPEN appears at the intersection of at least three seminal processes of eukaryotic transcription: chromatin remodeling, transcriptional initiation, and mRNA processing. These SPEN interactions also explain how it can act as either a coactivator or a corepressor.
To our knowledge, this is the first attempt to comprehensively investigate and address how diverse patterns of associations in HT-IP/MS datasets translate into a tiered organization of complex-complex interactions on a proteome scale. Next, we use transcriptional coregulators to show how this resource can be used to better understand existing, and to explore new, connections in transcriptional regulation.
Nuclear Receptor (NR) Transcriptional Regulation
The NURSA Consortium curates and maintains a tally of over 300 NR coregulators (Lanz et al., 2006); www.nursa.org; and Table S1). In our dataset, 128 NR coregulators were recovered as direct antigens (see Figure 1B for examples), while an additional 185 were identified as interacting proteins or in unintended cross-reacting complexes. We can confidently assign 170 MEMOs that contain 211 known NR coregulators and ~300 additional proteins that are likely coregulators. Furthermore, CCI networks of NR coregulators currently show >3,700 unique gene products that perticipate in coregulation. This is a considerable expansion of the NURSA coregulator list, suggesting that many more proteins can participate in the regulation of NR-driven transcription than previously thought.
Deconvolution of the CCI Network of SRC-3 Reveals A Distinct Interactive Subset of NR Coregulators
We noticed that many NR coregulators, although recovered at high numbers in multiple reciprocal IPs, did not consistently form preferred interactions. Such promiscuous interaction patterns were observed most often for multi-functional coregulators that perform extremely diverse cellular functions. We therefore broadly classify transcriptional coregulators into two classes according to the type of protein complexes they form.
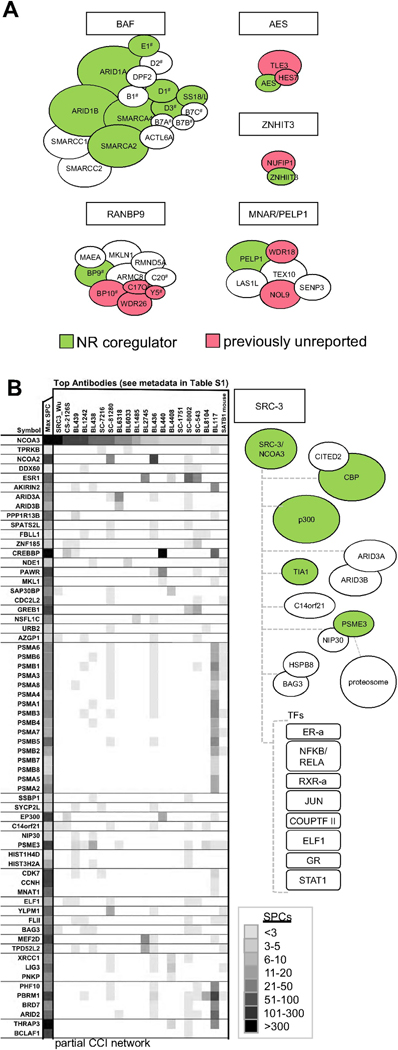
Type I coregulators exist in relatively stable multi-subunit steady-state complexes and show little variation in their composition, thus conforming to our conventional image of a protein complex. Examples of Type I complexes include well known Mediator (Gu et al., 1999), Nucleosome Remodeling and Deacetylase complexes (NURD) (Xue et al., 1998), CoREST complex (Hakimi et al., 2002), HDAC3/NCOR (Guenther et al., 2000), and SWI/SNF (BAF/P-BAF) (Kaeser et al., 2008; Wang et al., 1996), all of which we recovered fully in our IP/MS dataset, along with lesser known RANBP9, PELP1 (Figure 4A and S4A), and the Z3 complexes described below.
Figure 4. Coregulators Have Distinct Patterns of Protein Interaction Profiles.
3N analyses broadly classify coregulator networks into having either stable preferential protein networks (Type I) or multiple transient interactions (Type II). A) Schematic illustrations of examples of Type I NR coregulators with previously unidentified subunits. Green: coregulators listed at NURSA.org; E1, D2, D1, B1, and D3 are SMARC subunits; B7A,B,C are BCL7A,B, and C; C20, C20orf11; C17, C17orf39; BP9 and BP10 are RANBP9 and RANBP10; Y5, YPEL5, respectively. B) 3N heatmap excerpt for SRC-3/NCOA3. This is a typical Type II coactivator that lacks stoichiometric steady-state complex while revealing a multitude of sub-stoichiometric interactions, such as CBP (CREBBP), p300 (EP300), REG-gamma (PSME3), and various transcription factors (TFs).
The Type II coregulators, on the other hand, do not have consistent steady-state stoichiometric partners and often resolve into one-component MEMOs (Figure 4B and S4B). Such sub-stoichiometric associations likely represent the hallmark behavior for many coregulators, which is to provide a quick response to different cellular signals through dynamic context-specific associations. The oncogenic SRC-3 (a.k.a. AIB1 or NCOA3), is a prime example of a Type II coregulator.
The multifunctional transcriptional coregulator SRC-3 is a common target for cellular growth programs (Anzick et al., 1997; Kuang et al., 2004; Torres-Arzayus et al., 2004) (Lonard et al., 2007; Yan et al., 2006), adipogenic and energy balance (Louet et al., 2006); Coste A et al, 2008) and control of mRNA translation of pro-inflammatory cytokines (Yu et al., 2007). Such functional diversity may be provided by associated proteins, which bind SRC-3 proteins that are differentially modified post-translationally in response to the activation of different signaling pathways (Li et al., 2008; O'Malley et al., 2008; York et al., 2010). SRC-3 itself is a target of multiple cell signaling pathways (Font de Mora and Brown, 2000; Gianni et al., 2006; Wu et al., 2004; Yan et al., 2008; Yi et al., 2008), and acts as an integrator by converging signals to the genome for modulation of target gene expression. Our 3N analyses show only sub-stoichiometric interaction patterns in the SRC-3 interactome, even for well-proven associations such as CBP/p300 and REGγ (PSME3) (Figure 4B). Along with the interacting coregulators, a diverse number of NRs, such as ERα, RXRα/β and COUP-TFII, along with frequent tethering partners such as AP-1, co-precipitated with SRC-3 in our IP experiments. Importantly, this Type II behavior is preserved in MCF-7 breast cancer cells described in our earlier study (Lanz et al., 2010). These observations suggest that Type II behavior is an inherent and cell-type independent property of this molecule, and quite possibly also of many other Type II transcriptional coregulators.
The ability to distinguish Type I from Type II molecules serves as a good illustration of the conceptually different insights gained by a large-scale proteomics study, whose discovery-driven character is better positioned to reveal general perceptions, such as the overall biological functions of a Type II protein. A classification of transcriptional coregulators based on their ‘interactivity’ helped us to better understand why some coregulators have been reported to be responsible for many different cellular processes, while others seem to exert specialized biology only, albeit functioning in different signal and gene contexts. For Type II molecules, variability of interactants found in individual experiments for such proteins is overcome by analyzing large single IP/MS studies like the one presented here. It then becomes clear that a protein that biochemically appears not to form a consistent complex often displays the most complicated regulatory interaction networks, indicating its important functional significance at the CCI level rather than in the formation of core complexes. The identification of Type II interacting partners is indispensable for the understanding of regulatory processes, and will remain an important challenge for affinity-based approaches and for translational efforts. This is certainly the case for oncogenic SRC-3.
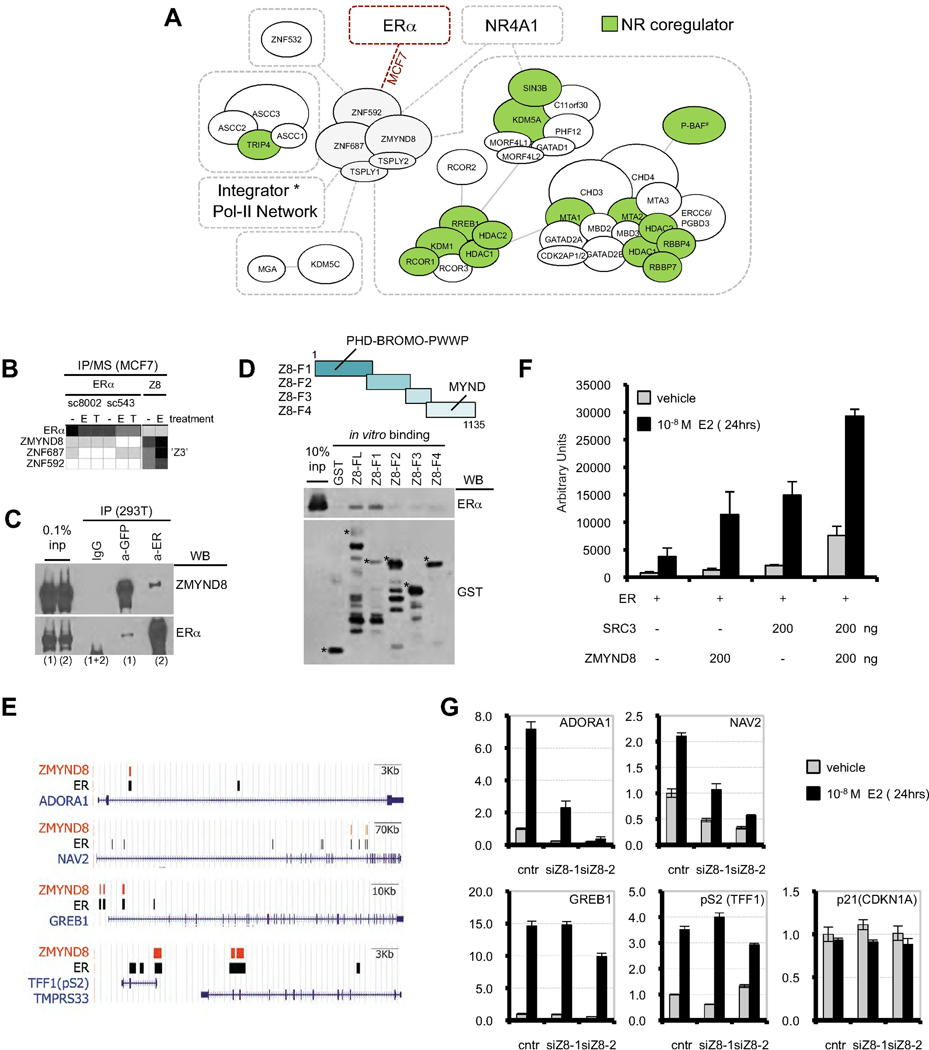
The Z3 Coregulator Complex Participates with an Extensive CCI Network
Among hitherto unspecified coregulators, our work revealed a transcriptional coregulator complex whose number of interactions rivals that of Mediator or Integrator (Fig.5A). This Z3 MEMO consists of ZMYND8, ZNF687, and ZNF592. ZMYND8 contains a PHD-finger, a Bromodomain, a Pro-Trp-Trp-Pro (PWWP) motif, a MYND-type zinc finger, and is likely to function as a reader of histone modifications. It recently has been suggested to affect chromatin silencing (Poleshko et al., 2010) and to interact with the transcriptional coregulator RCOR2 (Zeng et al., 2010). ZNF687 and ZNF592 have multiple C2H2 zinc fingers known to bind DNA.
Figure 5. Z3 Complex is a Transcriptional Coregulator.
A) Concise representation of the Z3 CCI network showing extensive interaction connections to transcriptional machinery (see also Figure S5A,B). Pol-II-Integrator network (*) is omitted due to space limitations. In addition to histone demethylases KDM5C, KDM5A and KDM1 (see text), Z3 also interacts with ASC1 (TRIP4) coregulator. (#) C112, C20orf112; P-BAF, Polybromo and Brg/Brahma-Associated Factor (see Figure 4A for BAF). Known NR coregulators are shown in green. B) Reciprocal IP/MS of Z3 proteins with ERa in MCF7 cells. C) Reciprocal IP/WB of overexpressed GFP-ZMYND8 and ERα in 293T cells; (1) and (2) are corresponding inputs. D) In vitro binding suggests that binding to ERα is via the N-terminal portion of ZMYND8 (Z8-F1). Asterisks indicate positions of ZMYND8 fragments. E) UCSC browser examples showing co-occupancy of ERα and ZMYND8 binding sites at E2-upregulated genes. F) ZMYND8 coactivates ERα in reporter luciferase assay. G) RNAi knockdown of ZMYND8 compromises upregulation of some E2-responsive target genes. p21 serves as a negative control.
Z3 forms a uniCORE with a two-protein MEMO consisting of TSPYL1/TSPYL2 (testis specific protein Y-like 1 and 2) (Figure 5A and S5A). The Integrator protein complex appears to be the predominant partner of Z3, bringing Pol-II into the Z3 CCI network. Z3 also is frequently found associated with the H3K4 demethylation machinery, including the H3K4me2/me1 demethylase KDM1 uniCORE, a partnering H3K4me3 demethylase KDM5A–containing SIN3B complex, and KDM5C H3K4me3 demethylase. Such conglomeration of histone modification readers suggests a prominent role for Z3 biology in interpreting the histone code for remodeling of chromatin for transcription. In addition, Z3 co-occurs with many transcription factors in our IP/MS dataset, including the nuclear receptor NR4A1 (NUR77, Figure S5B) and ERα, which we discuss next.
ERα interacts with the Z3 network at sub-stoichiometric levels (Figure 5B) in MCF7 breast cancer cells. We confirmed this by co-IP and in vitro GST pull-down assays (Figure 5C,D and S5C). Furthermore, ChIP-seq analysis showed a significant overlap of ZMYND8 chromatin binding sites with known ERα binding sites (Figure 5E and S5D). We also confirmed transcriptional coactivator functions of ZMYND8 in ERα-driven reporter assays and on endogenous E2-dependent genes (Figure 5F,G). siRNA knockdown of ZMYND8 showed markedly decreased transcription at the presumptive ERα/Z3 target genes ADORA1 and NAV2, while the classical ERα targets pS2/TFF1 and GREB1 appear to be less affected (Figure 5G), suggesting likely gene-specificity of ZMYND8. This example again demonstates the potential of our CCI resource to serve as a basis for deriving testable hypotheses.
Bridging the Gap Between Genome-Wide Data and Functional Proteomic Analyses
Currently, genome-wide association studies are identifying common genetic factors that impact health and disease. Even at an early stage the linking of genomics and proteomics data can further our understanding of disease pathways and provide platforms for new hypotheses. For example, in oncology it is imperative to distinguish causative driver mutations from ‘hitchhiking’ passengers (Stratton et al., 2009). Based on the premise that protein complexes function as biological units, we imply that proteins that physically associate with known cancer drivers ‘collectively’ confer a selective growth advantage on cells. These ‘guilt-by-association co-drivers’ thus appear as additional new valuable targets for functional translational analyses and therapeutic interventions. Cellular protein complexomes such as the resource presented here can complement genomic data with most valuable proteomics information, thus leading to a better understanding and treatments of human cancers.
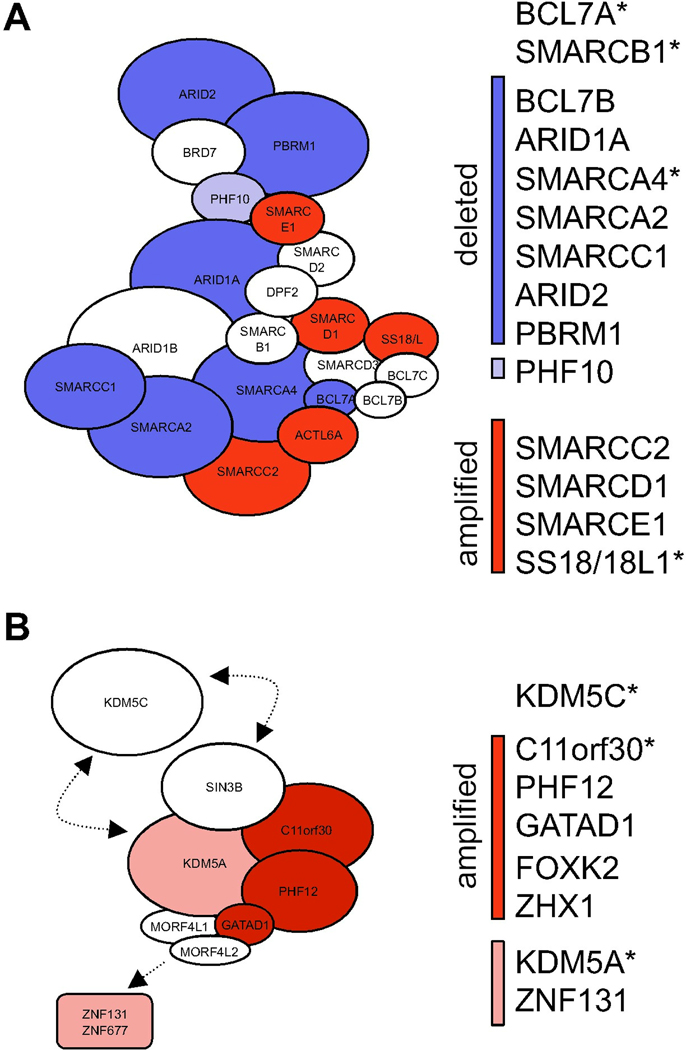
In Figure 6 we show two examples of the accrual of oncogenetic data in protein complexes. We used the Sanger Cancer Gene Census resource of human oncogenic mutations (Futreal et al., 2004) as well as the Broad Institute’s Tumorscape database, which lists copy number changes across multiple cancer types (Beroukhim et al., 2010) to flag cancer-related gene products. In the P-BAF uniCORE, for example, an overwhelming number of complex components have different associations with cancers (Figure 6A), strongly suggesting a key physiological role of this chromatin-remodeling complex as a whole in oncogenesis, and implying that its subunits are functionally more likely to be drivers rather than passengers.
Figure 6. Cancer Gene Alterations Group Within Protein Complex Modules.
A) The P-BAF complex is significantly perturbed in lung cancers. Red: amplified; dark blue: deleted in NSCLC; light blue: deleted in lung lineages. B) The SIN3B uniCORE is a hub of proteins with genomic amplifications. All but two proteins of the SIN3B complex are significantly amplified in breast cancer lineages (dark red) or epithelial lineages (pink). All genes marked with an asterisk are listed in Cancer Gene Sensus (Futreal et al.) and/or have been implicated causally in cancer development.
Similarly, the SIN3B uniCORE has three subunits that are significantly amplified in breast cancers (C11orf30, GATAD1, PHF12), while two more proximal components (KDM5A, ZNF131) are amplified in epithelial lineages (Figure 6B; (Beroukhim et al., 2010)). Among these, C11orf30, (EMSY) and KDM5A were identified as drivers in breast cancers (Brown et al., 2010; Raouf et al., 2005; Santarius et al., 2010) and in acute myeloid leukemia (Futreal et al., 2004; Glaros et al., 2007; Wang et al., 2009b), respectively. Although the other gene products in this complex have not yet been causally implicated in cancer, our finding that they reside in a stable complex with validated cancer drivers predicts that they also are likely to provide a selective advantage to clonal cell expansions.
Perspectives
Here, we present a large-scale HT-IP/MS dataset and discuss examples of extensive interaction networks derived from endogenous human regulatory protein complexes. Our approach serves as an operational framework for the deconvolution of high-content proteomics data, and testifies to the feasibility of mapping the endogenous protein complexome on a proteome-wide scale in higher eukaryotes.
The dense reciprocity of protein associations in our IP/MS dataset ensures high confidence assignments and provides the highest standard of verification for native protein-protein interactions. The analysis of multiple reciprocal immunoprecipitations forgoes the necessity to exhaustively characterize the specificity of primary antibodies and produces a low number of false positive designations.
We recovered most published core complexes, and have found many previously undocumented complex subunits. We also accomplished a characterization of weak protein interactions beyond most prior studies. As a result, we offer our hierarchical organization of endogenous protein complex interactions as a three-tiered organization of stable minimal endogenous modules (MEMOs), which combine to form variant core complexes (uniCOREs) that then interact with each other to form complex-complex interaction networks (CCIs). Our resource includes snapshots of high-order associations between regulatory complexes and serves as an initial roadmap for understanding the human interactome.
Because endogenous protein complexes were isolated and identified, our method is directly transferable to any cell line in culture, and will be extended to isolated tissues in the near future. This is particularly important when specificity for cell types and sensitivity to challenges such as hormonal treatments or exogenous stress must be considered to delineate particular context-dependent interactions. Ultimately, mapping the context-dependent interactomes is a molecular requisite for understanding cellular biology.
This study emphasized nuclear transcriptional coregulators and in Type I and Type II complexes revealed two broadly distinct coregulator interactive patterns. Further, we argue that when intelligently combined with genomics and transcriptomics data, proteomics information provides a rationale and predictable understanding of phenotype that has a real potential for translation. Our recurrent finding that cancer gene products group together in select protein complexes supports the idea that the perturbation of a protein complex as a whole should be implicated in the etiology of malignancies, entailing all complex constituents as valuable molecular targets for disease screening and therapy. This same logic can be applied to the etiology of polygenic metabolic or CNS diseases, where mutations in multiple genes are responsible for a disorder. We postulate that in many cases, polygenic mutations compromise the physiologic function within a single multi-protein complex, and then additional mutations further degrade the function of the particular interaction network above the threshold for disease progression. In this manner, our resource will provide ample ideas for further testing of the involvement of coregulatory proteins in the development of human polygenic diseases.
Taken together, genomics, proteomics, and targeted functional studies comprise three critical components of the combinatorial ‘systems’ approach that can be employed to effectively translate genomics data into molecular knowledge in order to ultimately advance clinical applications.
Experimental Procedures
Cell Culture and Nuclear Extraction
HeLa S3 were cultured in suspension in 20–36L spin bottles in RPMI-1640 media with 5% FBS to final density of 0.5×106 cells/ml. Attached cell lines were grown in 50–200 15cm plates in DMEM with 5% FBS. Cell fractionation was done as previously described (Malovannaya et al., 2010). For HeLa S3, nuclear extraction with 0.02–1.2M KCl buffer titration was applied; for MCF7 cells, the 300–900 mM KCl gradient was used. Resulting ~500 mM salt extract was dialyzed to 160 mM KCl.
Immunoprecipitation
IPs were carried out essentially as described (Malovannaya et al., 2010). Briefly, nuclear extracts were ultra-centrifuged (200,000 rcf) after thawing; 0.5– 1 ml of extract (~10–15mg) was incubated with 7–15µg of primary antibody for 2h followed by ultra-centrifugation and 45 min incubation with Sepharose-CL4B Protein A beads (GE Healthcare). Minimal bead washing with NTN (50mM Tris-Cl pH8.0, 150mM NaCl, 0.5% NP-40) was used to preserve transient interactions. Information about intended and predicted antigens of particular antibodies can be found in Table S1.
SDS-PAGE and Mass Spectrometry
Immunocomplexes were eluted in 1X Laemmli buffer and resolved on pre-cast 4–20% Novex Tris-Glycine or 4–12% NuPage gels (Invitrogen, CA) to half-length. Gels were minimally stained with Coomassie Brilliant Blue, cut into 6 molecular weight ranges and heavy chain IgG band, and digested with trypsin. Immunocomplexes were identified on a Thermo Fisher LTQ (majority) or Velos-Orbitrap mass spectrometers. Spectral data were then searched against human protein RefSeq database in BioWorks or Proteome Discoverer Suites with either SeQuest (for LTQ data) or Mascot (Orbitrap data) software. Multi-consensus result files of protein GI identifiers were compiled for each IP with strict filters and often manually inspected.
Data Processing
IP/MS results were transferred into an in-house built FileMaker-based relational database where protein GIs were converted to the GeneID identifiers according to the NCBI ‘gene2accession’ table. Data filtering and deconvolution was performed as described in the text and in Extended Experimental Procedures.
Overexpression and reciprocal co-IP
293T cells were transiently transfected with expression constructs for estrogen receptor (pCR3.1-hERα), pEGFP-C1-ZMYND8 (inserted at BglII/EcoRI sites), or control plasmids using Lipofectamine 2000 (Invitrogen). 48h after transfection, cells were lysed in BC lysis buffer (20 mM Tris-HCl pH7.5, 0.2 mM EDTA, 20% glycerol, 0.15 M NaCl, 0.5% NP40, 10 mM β-ME and PIs), lyzates were centrifuged at 100,000 rcf and immunoprecipitated using 1 µg anti-ER (SC-8002, SCBT), anti-GFP (custom, Genemed Synthesis), or control rabbit/mouse IgG mix (Cell Signaling), followed by SDS-PAGE and Western blotting.
RNA interference and real-time quantitative PCR
3×105 MCF7 cells were transfected with 100 pmoles siRNA against ZMYND8 (HSS119059 and 1199061) or Negative Control Medium CG Content #2 siRNA (Invitrogen) using Lipofectamine RNAiMAX reverse transfection protocol. Cells were changed to 5% stripped serum DMEM media w/o phenol red at 16h, and at 36h treated with 10 nM β-estradiol (Sigma) and collected 24h after treatment. Total RNA was isolated using RNeasy kit (QIAGEN); cDNA was prepared using SSIII RT system (Invitrogen) and analyzed by quantitative RT-PCR on ABI 7500 or StepOne Plus thermocyclers (Applied Biosystems) using SYBR Green (Applied Biosystems/LifeTechnologies) and gene-specific primers (see Figure S5D).
Luciferase reporter assays
HeLa cells were transfected with pCR3.1-hERα, pERE-E1b–LUC, pCMV-Tag2-SRC3, pCRIITOPO-ZMYND8, and/or control vectors using Lipofectamine 2000 (Invitrogen) and cultured in 5% stripped serum-containing DMEM w/o phenol red. 48h after transfection, cells were treated overnight with 10 nM β-estradiol or ethanol vehicle, lysed in luciferase lysis buffer (25 mM Tris-HCl pH8.0, 150 mM NaCl, 0.5% Triton-X100, 10% grycerol, 1 mM EDTA, 1 mM DTT, PIs), and luciferase was measured using Promega luciferase assay system on Centro LB960 luminometer (Berthold Technologies).
GST pull-down
GST fusion proteins (Figure S5C) were produced in BL21 bacteria (Stratagene) using 0.5 mM IPTG induction. Bacterial pellets were lysed in NETN buffer (50 mM Tris-HCl pH8.0, 150 mM NaCl, 0.5% NP40, 5 mM EDTA, PIs), cleared by ultracentrifugation and bound to glutathione sepharose 4B (GE Healthcare) for 1 hr at 4°C. Beads were washed with NETN, and amount of GST protein was estimated by SDS-PAGE followed by Coomassie staining. GST fusion proteins or control GST bound to beads were pre-incubated with 100 µl NETN containing 10 µg/ml BSA for 1 hr at 4°C to block non-specific binding. Recombinant estrogen receptor was obtained from Invitrogen, diluted in NETN to 0.2 µg/ml and cleared by ultracentrifugation. 100 µl of ER solution was added to 100 µl of bead-bound domains and incubated for 20 min at 4°C, followed by triple NETN washes. Pull-downs were analyzed by Western blotting.
Genome-wide ZMYND8 location analysis by ChIP-sequencing
Chromatin immunoprecipitation was performed according to the protocol from Upstate Biotechnology with minor modifications. 1×10^7 MCF7 cells were used for each ChIP after 45min treatment with 10 nM β-estradiol. After sonication the chromatin fraction was immunoprecipitated with CBC2032 ZMYND8 antibody and freed by heating. The ChIP-seq data were processed using MACS 1.3.5 with p-values 1e8 and fold-change greater than 40 for peak calls.
Highlights.
- a database of endogenous coregulator complexes containing >10000 unique proteins
- a data analysis approach apt for weaker complex-complex interaction networks
- many transcriptional coregulators promiscuously share multiple interaction networks
- complex is a functional unit that can explain diverse etiologies of polygenic disease
Supplementary Material
01
02
03
Acknowledgements
This work is supported partially by the NURSA grant U19-DK62434 including the Proteomics Strand funding to B.W.O. and J.Q. and the Collaborative Bridging Project funding to R.B.L. We acknowledge the significant financial support of the Center for Molecular Discovery and the McLean Foundation at Baylor College of Medicine. NIH grants HD08188 and DK059820 to B.W.O., CA84199 to J.Q., GM080703 to Y.W., and CPRIT RP101499 fellowship to N.Y. are also acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
CCI networks of nuclear receptor coregulators have been incorporated into NURSA molecular pages and standalone resource solutions for Mac and PC are available for downloading at the NURSA website (www.nursa.org).
References
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LA, Johnson K, Leung S, Bismar TA, Benitez J, Foulkes WD, Huntsman DG. Co-amplification of CCND1 and EMSY is associated with an adverse outcome in ER-positive tamoxifen-treated breast cancers. Breast Cancer Res Treat. 2010;121:347–354. doi: 10.1007/s10549-009-0479-x. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem. 2010;285:4268–4272. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–728. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font de Mora J, Brown M. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol Cell Biol. 2000;20:5041–5047. doi: 10.1128/mcb.20.14.5041-5047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gianni M, Parrella E, Raska I, Jr, Gaillard E, Nigro EA, Gaudon C, Garattini E, Rochette-Egly C. P38MAPK-dependent phosphorylation and degradation of SRC-3/AIB1 and RARalpha-mediated transcription. EMBO J. 2006;25:739–751. doi: 10.1038/sj.emboj.7600981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Glaros S, Cirrincione GM, Muchardt C, Kleer CG, Michael CW, Reisman D. The reversible epigenetic silencing of BRM: implications for clinical targeted therapy. Oncogene. 2007;26:7058–7066. doi: 10.1038/sj.onc.1210514. [DOI] [PubMed] [Google Scholar]
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci U S A. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Tachikawa H, Miura Y, Takahashi N. TRIM11 binds to and destabilizes a key component of the activator-mediated cofactor complex (ARC105) through the ubiquitin-proteasome system. FEBS Lett. 2006;580:4784–4792. doi: 10.1016/j.febslet.2006.07.066. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser MD, Aslanian A, Dong MQ, Yates JR, 3rd, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Chen J. New players in the BRCA1-mediated DNA damage responsive pathway. Mol Cells. 2008;25:457–461. [PMC free article] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher T, Superti-Furga G. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat Methods. 2007;4:807–815. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kuang SQ, Liao L, Zhang H, Lee AV, O'Malley BW, Xu J. AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res. 2004;64:1875–1885. doi: 10.1158/0008-5472.can-03-3745. [DOI] [PubMed] [Google Scholar]
- Lanz RB, Bulynko Y, Malovannaya A, Labhart P, Wang L, Li W, Qin J, Harper M, O'Malley BW. Global characterization of transcriptional impact of the SRC-3 coregulator. Mol Endocrinol. 2010;24:859–872. doi: 10.1210/me.2009-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz RB, Jericevic Z, Zuercher WJ, Watkins C, Steffen DL, Margolis R, McKenna NJ. Nuclear Receptor Signaling Atlas ( www.nursa.org): hyperlinking the nuclear receptor signaling community. Nucleic Acids Res. 2006;34:D221–D226. doi: 10.1093/nar/gkj029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Liang YY, Feng XH, Tsai SY, Tsai MJ, O'Malley BW. Essential phosphatases and a phospho-degron are critical for regulation of SRC-3/AIB1 coactivator function and turnover. Mol Cell. 2008;31:835–849. doi: 10.1016/j.molcel.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, Lanz RB, O'Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Louet JF, Coste A, Amazit L, Tannour-Louet M, Wu RC, Tsai SY, Tsai MJ, Auwerx J, O'Malley BW. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proc Natl Acad Sci U S A. 2006;103:17868–17873. doi: 10.1073/pnas.0608711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Li Y, Bulynko Y, Jung SY, Wang Y, Lanz RB, O'Malley BW, Qin J. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci U S A. 2010;107:2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- O'Malley BW, Qin J, Lanz RB. Cracking the coregulator codes. Curr Opin Cell Biol. 2008;20:310–315. doi: 10.1016/j.ceb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleshko A, Einarson MB, Shalginskikh N, Zhang R, Adams PD, Skalka AM, Katz RA. Identification of a functional network of human epigenetic silencing factors. J Biol Chem. 2010;285:422–433. doi: 10.1074/jbc.M109.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf A, Brown L, Vrcelj N, To K, Kwok W, Huntsman D, Eaves CJ. Genomic instability of human mammary epithelial cells overexpressing a truncated form of EMSY. J Natl Cancer Inst. 2005;97:1302–1306. doi: 10.1093/jnci/dji254. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Res. 2010;38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci U S A. 2009a;106:3166–3171. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, Greenberg RA. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009b;23:740–754. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Arzayus MI, Font de Mora J, Yuan J, Vazquez F, Bronson R, Rue M, Sellers WR, Brown M. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–274. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009a;23:729–739. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009b;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O'Malley BW. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Yan J, Erdem H, Li R, Cai Y, Ayala G, Ittmann M, Yu-Lee LY, Tsai SY, Tsai MJ. Steroid receptor coactivator-3/AIB1 promotes cell migration and invasiveness through focal adhesion turnover and matrix metalloproteinase expression. Cancer Res. 2008;68:5460–5468. doi: 10.1158/0008-5472.CAN-08-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, Jetten AM. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007;67:6647–6656. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Tsai SY, Tsai MJ. SRC-3/AIB1: transcriptional coactivator in oncogenesis. Acta Pharmacol Sin. 2006;27:387–394. doi: 10.1111/j.1745-7254.2006.00315.x. [DOI] [PubMed] [Google Scholar]
- Yi P, Feng Q, Amazit L, Lonard DM, Tsai SY, Tsai MJ, O'Malley BW. Atypical protein kinase C regulates dual pathways for degradation of the oncogenic coactivator SRC-3/AIB1. Mol Cell. 2008;29:465–476. doi: 10.1016/j.molcel.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York B, Yu C, Sagen JV, Liu Z, Nikolai BC, Wu RC, Finegold M, Xu J, O'Malley BW. Reprogramming the posttranslational code of SRC-3 confers a switch in mammalian systems biology. Proc Natl Acad Sci U S A. 2010;107:11122–11127. doi: 10.1073/pnas.1005262107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, York B, Wang S, Feng Q, Xu J, O'Malley BW. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell. 2007;25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Kong Q, Li C, Mao B. Xenopus RCOR2 (REST corepressor 2) interacts with ZMYND8, which is involved in neural differentiation. Biochem Biophys Res Commun. 2010;394:1024–1029. doi: 10.1016/j.bbrc.2010.03.115. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03