Single-cell dissection of transcriptional heterogeneity in human colon tumors (original) (raw)
. Author manuscript; available in PMC: 2012 Jun 1.
Published in final edited form as: Nat Biotechnol. 2011 Nov 13;29(12):1120–1127. doi: 10.1038/nbt.2038
Abstract
Cancer is often viewed as a caricature of normal developmental processes, but the extent by which its cellular heterogeneity truly recapitulates multi-lineage differentiation processes of normal tissues remains unknown. Here, we implement “single-cell PCR gene-expression analysis” (SINCE-PCR) to dissect the cellular composition of primary human normal colon and colon cancer epithelia. We show that human colon cancer tissues contain distinct cell populations whose transcriptional identities mirror those of the different cellular lineages of normal colon. By creating monoclonal tumor xenografts from injection of a single-cell (n = 1), we show that transcriptional diversity of cancer tissues is largely explained by in vivo multi-lineage differentiation, not only by clonal genetic heterogeneity. Finally, we show that perturbations in gene-expression programs linked to multi-lineage differentiation strongly associate with patient survival. Guided by SINCE-PCR data, we develop two-gene classifier systems (KRT20 vs CA1, MS4A12, CD177, SLC26A3) that predict clinical outcomes with hazard-ratios superior to pathological grade and comparable to microarray-derived multi-gene expression signatures.
The in vivo cellular composition of solid tissues is often difficult to investigate in a comprehensive and quantitative way. Techniques such as immunohistochemistry and flow cytometry are limited by the availability of antigen-specific monoclonal antibodies and by the small number of parallel measurements that can be performed on each individual cell. Traditional high-throughput assays, such as gene-expression arrays, when performed on whole tissues, provide information on average gene expression levels, and can be only indirectly correlated to quantitative modifications in cellular subpopulations. These limitations become particularly difficult to overcome when studying minority populations, such as stem cells, whose identification is made elusive by their low numbers and by the lack of exclusive markers. Moreover, in pathological states, such as cancer, it is frequently impossible to determine whether perturbations in gene expression detected in whole tissues are due to modifications in the relative composition of different cell types or to aberrations in the gene-expression profile of mutated cells.
For example, although it has been postulated that multi-lineage differentiation can contribute to tumor heterogeneity 1–3, this issue remains controversial 4. Many in the field argue that heterogeneity is mainly the result of clonal evolution as a result of genomic instability 5, 6. Previous studies addressed this question, but could rely only on in vitro cultured cell lines and on simple morphological evidence 7–9. Moreover, recent evidence indicates that, in the absence of a molecular proof of monoclonal origin, results from in vitro experiments based on limiting dilution can be biased due to a dramatic increase in cell survival by cell hetero-doublets. This phenomenon is best exemplified in the case of the mouse small intestine, where growth and expansion of LGR5+ progenitor cells is dramatically enhanced by the co-presence of a bystander epithelial feeder cell 10. Based on these studies, it remained difficult to perform a quantitative measure of the extent of multi-lineage differentiation in cancer tissues and, above all, to investigate to what extent it actually translated into the differential activation of distinct transcriptional programs that would mirror and recapitulate the physiological processes observed in normal tissues.
RESULTS
Description and technical validation of the SINCE-PCR method
We combined “fluorescence activated cell sorting” (FACS) and “single-cell PCR gene-expression analysis” (SINCE-PCR) to perform a high-throughput transcriptional analysis of the distinct cellular populations contained in solid human tissues (Supplementary Fig. 1 and 2). This method exploits the capacity of modern flow cytometers to sort individual single cells with accuracy and precision (Supplementary Fig. 3), together with the use of microfluidic technologies to perform high sensitivity multiplexed PCR from minute amounts of mRNA, thereby allowing parallel analysis of the expression of up to 96 genes for each individual cell. The large number of measurements per cell and the possibility to analyze several hundreds of cells in parallel from the same sample, allow the use of statistical clustering algorithms in order to associate cells with similar gene expression profiles into well defined subpopulations (Supplementary Fig. 2). Microfluidic platforms have been previously validated for single-cell gene-expression analysis 11–13. Consistent with those results, our control experiments with titrated mRNA standards as well as single-cell experiments on a cell line validated the sensitivity of this approach for high throughput analysis across multiple genes (Supplementary Fig. 4).
SINCE-PCR analysis of normal human colon epithelium: discovery of novel markers and novel cell populations
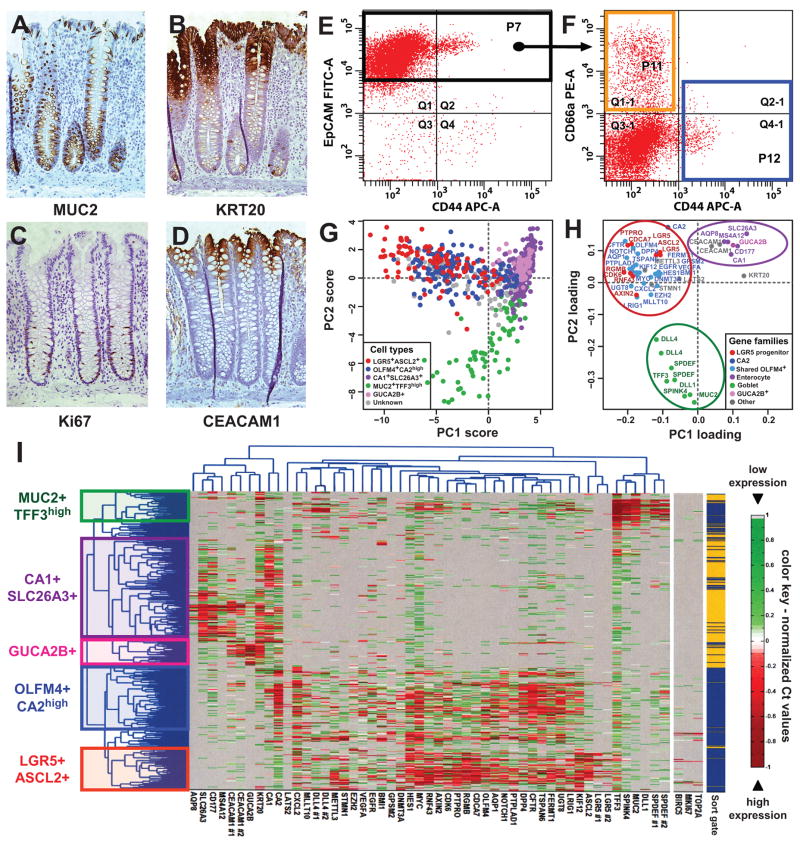
We first applied SINCE-PCR to the study of normal human colon epithelial cells. Human colon epithelium is composed of heterogeneous populations of cells which express different protein markers based on their lineage, differentiation stage and functional status. Many of these cell subsets can be identified by immunohistochemistry against well characterized markers, such as MUC2, which encodes for a mucin glycoprotein expressed by goblet cells, KRT20, which encodes for an intermediate filament protein preferentially expressed by differentiated colon epithelial cells, and Ki67, which is expressed by proliferating cells (Fig. 1, A–C) 14. In normal conditions, immature colon epithelial cells reside at the bottom of the colonic crypts and express high levels of the surface marker CD44, while differentiated mature cells progressively migrate to the top and progressively lose CD44 expression 14, 15. We focused our analysis on the stem/immature compartment of the colonic epithelium by sorting the EpCAMhigh/CD44+ population (Fig. 1, E–F), which, in normal tissues, corresponds to the bottom of the human colonic crypt 14. To study the more mature, terminally differentiated cell populations, we analyzed an equal number of cells from the EpCAM+/CD44neg/CD66ahigh population, which corresponds to the top of the human colonic crypt (Fig. 1, D, F) 16. In our first pilot experiments, we tested the method’s feasibility using well established reference markers. We analyzed and clustered colon epithelial cells using three genes encoding for markers linked to either one of the two major cell lineages (i.e. MUC2 for goblet cells and CA1 for enterocytes) or the immature compartment (i.e. LGR5) of the colon epithelium 14, 17–19. This experiment showed that genes encoding for lineage-specific markers are frequently expressed in a mutually exclusive way, mirroring the expression pattern of corresponding proteins (Supplementary Fig. 5).
Figure 1. SINCE-PCR analysis of human normal colon epithelium.
Human colon epithelium is composed by a heterogeneous population of cells, which differ in expression of multiple markers, based on their lineage (A, goblet cells express MUC2), differentiation stage (B, “top-of-the-crypt” cells express high levels of KRT20) and proliferative status (C, proliferating cells express Ki67). In the normal colon epithelium, “top-of-the-crypt” and “bottom-of-the-crypt” epithelial cells can be differentially enriched by flow cytometry based of the expression of EpCAM, CD44 and CD66a/CEACAM1 (D–F). “Bottom of the crypt” epithelial cells were defined as EpCAM+/CD44+ (F, P12 blue sort gate) and “top-of-the-crypt” epithelial cells as EpCAM+/CD44neg/CD66ahigh (F, P11 orange sort gate). SINCE-PCR analysis of “top-of-the-crypt” and “bottom-of-the-crypt” normal colon epithelial cells led to the discovery of novel lineage and/or differentiation markers and the establishment of a core set of 57 TaqMan assays that allow the visualization of distinct cell populations, including enterocyte-like cells (CA1+/SLC26A3+ and GUCA2B+), goblet-like cells (MUC2+/TFF3high) and two compartments defined by gene-expression profiles reminiscent of more immature progenitors (OLFM4+/CA2high and LGR5+/ASCL2+) (I). CA1+/SLC26A3+ and GUCA2B+ cells were preferentially observed in the EpCAM+/CD44neg/CD66ahigh population (P11 orange sort gate), while MUC2+/TFF3high, OLFM4+/CA2high and LGR5+/ASCL2+ cells were preferentially observed in the EpCAM+/CD44+ population (P12 blue sort gate). Principal component analysis (PCA) of SINCE-PCR data confirmed hierarchical clustering results, visualizing distinct cell populations characterized by the coordinated expression of independent sets of genes (G–H; PC1: principal component #1, PC2: principal component #2). Both cell populations and gene families identified by PCA closely mirrored those identified by hierarchical clustering.
We then searched for novel gene-expression markers of the different cell populations, with a special focus on putative stem cell markers. We performed a high-throughput screening of 1568 publicly available gene-expression array datasets from human colon epithelia (Supplementary Table 1), using a bioinformatics approach designed to identify developmentally regulated genes based on Boolean implication logic (Supplementary Fig. 6) 20. The search yielded candidate genes whose expression associated with that of other markers previously linked to individual colon epithelial cell lineages (Supplementary Fig. 7–9). Using an iterative approach, we screened by SINCE-PCR more than 230 genes on 8 independent samples of normal human colon epithelium. At each round, genes that were non-informative (i.e. not differentially expressed in either positive or negative association with CA1, MUC2 or LGR5) were removed and replaced with new candidate genes. Thereby, we progressively built a list of 57 TaqMan assays that allowed us to analyze the expression pattern of 53 distinct genes (Supplementary Table 2) with high robustness (Supplementary Fig. 10). This allowed us to visualize and characterize multiple cell populations, using both hierarchical clustering (Fig. 1, I) and principal component analysis (PCA; Fig 1, G–H).
Analysis of the EpCAMhigh/CD44neg/CD66ahigh population (enriched for “top-of-the-crypt” cells) revealed that this subset, although transcriptionally heterogeneous, was almost exclusively composed of cells expressing high-levels of genes characteristic of mature enterocytes (e.g. CA1+, CA2+, KRT20+, SLC26A3+, AQP8+, MS4A12+) 14, 21–23 and led to the discovery of at least two novel differentially expressed gene expression markers (e.g. CD177, GUCA2B) (Fig. 1, H). To validate the reliability of SINCE-PCR results, we evaluated the distribution of SLC26A3 and CD177 protein expression in tissue sections and we confirmed its preferential expression at the top of the human colonic crypts (Supplementary Fig. 11 and 12). At the present time, it is not clear whether the different cell subsets observed within this population (e.g. CA1+/SLC26A3+ vs GUCA2B+) represent distinct stages of differentiation or distinct functional subsets of colonic enterocytes. Nonetheless, their clearly unique transcriptional programs identify them as part of a distinct cellular population.
Analysis of the EpCAMhigh/CD44+ population (enriched for “bottom-of-the-crypt” cells) revealed the presence of multiple populations, including: a) a cell compartment characterized by the expression of genes linked to goblet cells (MUC2+, TFF3high, SPDEF+, SPINK4+) 24, 25, b) a cell compartment characterized by the co-expression of genes associated to immature cells as well as genes known to be expressed by enterocytes (OLFM4+, CA2high) and c) a cell compartment whose gene-expression profile mirrors that of a stem/progenitor cell compartment in the mouse small intestine (LGR5+, ASCL2+, PTPRO+, RGMB+) 17, 26. A synopsis of the key genes that define the gene-expression profile of the different populations is provided in Supplementary Table 3.
The OLMF4+/CA2high and the LGR5+/ASCL2+ compartments shared expression of several genes of functional interest in both stem cell and cancer biology, such as genes involved in self-renewal and chromatin remodeling (EZH2, BMI1) 27–29, Wnt-pathway signaling (AXIN2)30, cell growth and chemotaxis (CXCL2)31, stem cell quiescence (LRIG1)32 and oncogenes (MYC)33. Of particular interest was also the gene-expression pattern of proliferation markers (i.e. MKI67, TOP2A, BIRC5/Survivin), whose expression appeared restricted to the EpCAMhigh/CD44+ (“bottom-of-the-crypt”) population, and particularly enriched in LGR5+/ASCL2+ and MUC2+/TFF3high cells, as partially expected based both previously published data 14, 17, 19 and our own immunohistochemistry results (Supplementary Fig. 13, C).
Among the novel findings obtained by SINCE-PCR is the observation that MUC2+/TFF3high cells are characterized by high-levels of expression of several genes of interest, including DLL1, DLL4 and KRT20. At first, the expression of KRT20 in the bottom of the crypt appeared contrary to the notion of KRT20 as a terminal differentiation marker. However, upon more careful examination of immunohistochemical stainings, we were able to clearly identify scattered KRT20+ cells, which can be morphologically identified as goblet cells (Supplementary Fig. 13, A–B). We also noticed that MUC2+/TFF3high cells, for the most part, lack expression of CFTR. The differential expression of DLL4 is of potential relevance to the clinical development of novel anti-tumor therapeutic agents 34.
SINCE-PCR analysis of a primary human colon adenoma
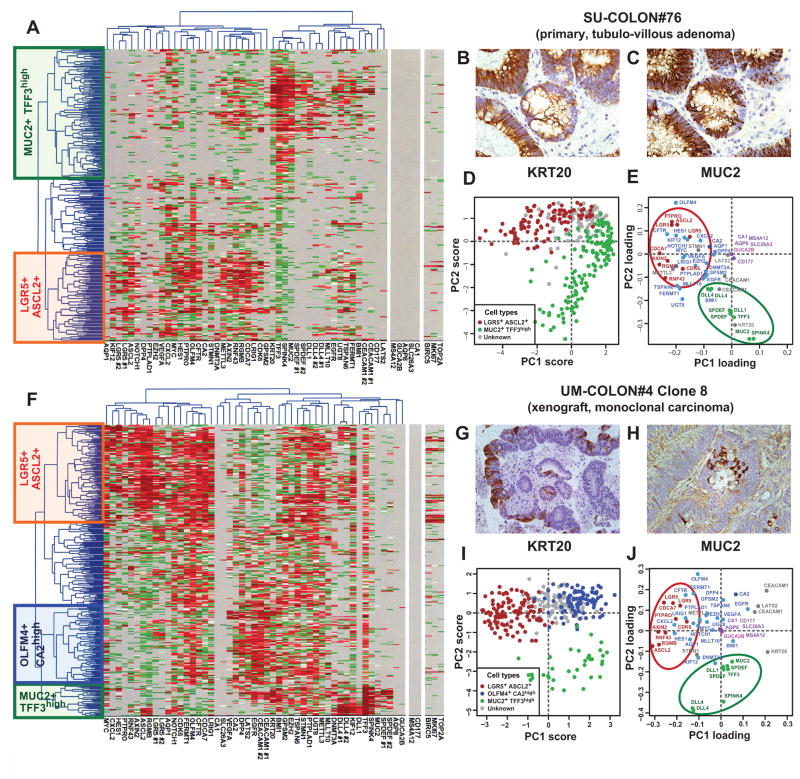
We then turned to cancer and investigated whether the cellular composition of the normal colonic epithelium is preserved in colorectal tumors, both benign and malignant. Analysis by SINCE-PCR of EpCAMhigh/CD44+ cells from a primary tubulo-villous adenoma (SU-COLON#76) revealed the presence of at least two different cell populations (i.e. LGR5+/ASCL2+ and MUC2+/TFF3high) characterized by distinctive gene signatures, closely mirroring those observed in corresponding EpCAMhigh/CD44+ populations of normal tissues (Fig. 2, A, D–E). These observations were confirmed at the protein level by parallel immunohistochemical investigations for KRT20 and MUC2 (Fig 2, B–C) and are in agreement with the recent notion that KRT20 is frequently expressed in opposition to LGR5 19. Interestingly, this primary adenoma appeared depleted in CA1+/SLC26A3+, GUCA2B+and OLFM4+/CA2high cell populations. Although at first unexpected, a careful examination of public gene-expression array databases indicated that this anomaly is likely to be a common feature of many benign adenomas (Supplementary Fig. 14).
Figure 2. SINCE-PCR analysis of human colon tumor tissues.
Analysis by SINCE-PCR of the EpCAM+/CD44+ population from human colon tumor tissues was performed on a large primary benign adenoma (A, SU-COLON#76) and a monoclonal colon cancer xenograft obtained from injection of a single-cell (n = 1) in a NOD/SCID/IL2Rγ−/− mouse (F, UM-COLON#4 Clone 8). The analysis revealed the presence of multiple cell populations characterized by distinct gene signatures, closely mirroring lineages and differentiation stages observed in the EpCAM+/CD44+ population from the normal colon epithelium. Principal component analysis (PCA) of SINCE-PCR data confirmed hierarchical clustering results, visualizing distinct cell populations characterized by the coordinated expression of independent sets of genes that corresponded to those observed in normal tissues (D–E, SU-COLON#76; I–J, UM-COLON#4 Clone8). Most importantly, the analysis of the monoclonal tumor xenograft obtained from a single-cell indicated that these distinct cell populations, together with their distinctive gene-expression repertoires, did not arise as the result of the coexistence within the tumor tissue of independent genetic sub-clones, but as the result of multi-lineage differentiation processes during tumor growth. SINCE-PCR data were confirmed at the protein level by immunohistochemistry, testing for expression of KRT20 and MUC2 on corresponding tissue sections (B–C, SU-COLON#76; G–H, UM-COLON#4 Clone 8). Color coding of normalized Ct values in hierarchical clustering plots (A, F) and of gene-families in PC loading plots (E–J) are identical to Fig. 1.
SINCE-PCR analysis of a human colon cancer xenograft derived from a single cancer cell
Tumor tissues, both benign and malignant, are known to undergo perturbations of normal differentiation processes, but it’s unclear to what extent those perturbations reflect quantitative changes in cell composition or qualitative changes in gene-expression programs. This topic has historically remained extremely controversial 4–9, 35. Our own systematic study of KRT20 and MUC2 protein expression in human malignant colorectal cancer tissues, for instance, revealed that both markers are frequently expressed heterogeneously, in patterns that mirror those observed in normal colorectal epithelium (Supplementary Fig. 15). It remained unclear, however, to what extent cancer transcriptional heterogeneity is the result of clonal genetic heterogeneity 36 or epigenetic heterogeneity due multi-lineage differentiation processes 9.
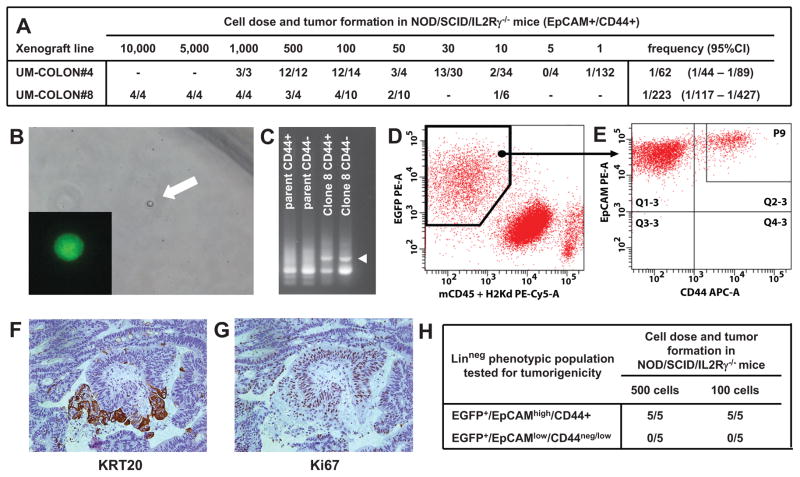
To address this question from a functional perspective, we investigated whether a single (n = 1) human colorectal cancer cell can recreate the heterogeneous cell composition of parent tumor tissues, including the subpopulations that we discovered in this study. We created tumors that originated from injection of a single (n = 1) EpCAMhigh/CD44+ cancer cell purified from one of our well-characterized solid xenograft lines 37, following infection with a lentivirus vector encoding for enhanced green fluorescence protein (EGFP; Fig. 3, A–B). Monoclonal origin of the tumors was confirmed by identification of a unique lentivirus integration site in all cancer cells (Fig. 3, C). Strikingly, the single-cell derived, lentivirus-tagged, EGFP+ tumors closely reproduced the phenotypic diversity of their parent tumors, both in terms of tissue histology and surface-marker phenotypic repertoire of cellular populations (Fig. 2, G–H; Fig. 3, D–G). Tumorigenicity experiments performed in NOD/SCID/IL2Rγ−/− mice revealed that, as observed in the parent tumors 37, EGFP+/EpCAMhigh/CD44+ and EGFP+/EpCAMlow/CD44neg/low cell populations were endowed with different tumorigenic capacity (Fig. 3, H). Most interestingly, a SINCE-PCR analysis of the EpCAMhigh/CD44+ population from these monoclonal tumors demonstrated its heterogeneous lineage composition, showing the presence of three distinct compartments (i.e. LGR5+/ASCL2+, OLFM4+/CA2high, MUC2+/TFF3high), again characterized by distinctive gene signatures, closely mirroring those observed in corresponding immature populations of normal tissues (Fig. 2, F, I–J). Taken together, these data formally prove that, at least in a subset of tumors, transcriptional heterogeneity is, at least partly, explained by multi-lineage differentiation processes that tend to recapitulate those observed in normal tissues.
Figure 3. Analysis of a monoclonal human colon cancer xenograft obtained from injection of a single-cell (n =1) in NOD/SCID/IL2Rγ−/− mice.
In human colon cancer, tumorigenic capacity in immunodeficient mice is enriched in a phenotypic subset of epithelial cancer cells (EpCAMhigh/CD44+). Within the EpCAMhigh/CD44+ population, the number of cells needed to establish a tumor varies based on the individual xenograft line (A), although tumor formation can be obtained even upon injection of very small numbers of cells, including single-cells (B). Monoclonal tumors derived from injection of a single (n = 1), lentivirus-tagged, EGFP+/EpCAMhigh/CD44+ cancer cell from human colon cancer xenograft UM-COLON#4 (B) bear unique lentivirus integration sites as compared to their polyclonal parent tumors (C) and reproduce the diversity of parent tumors both in terms of the phenotypic repertoire of cell populations (D–E) and tissue histology (F, KRT20; G, Ki67). Similar to what observed in parent tumors, EpCAMhigh/CD44+ and EpCAMlow/CD44neg/low populations are characterized by different tumorigenic capacity, as evaluated by tumorigenicity experiments in NOD/SCID/IL2Rγ−/−mice (H).
Prognostic implications of biomarkers identified by SINCE-PCR
To gain further insight in the potential functional implications of these observations, we then performed a comparative analysis of SINCE-PCR expression data in relation to genes associated with cell proliferation (i.e. Ki67, TOP2A, BIRC5/Survivin). In this case, too, we observed that their expression pattern in malignant tissues frequently mirrored that of normal ones. Both in the normal tissue and in the monoclonal human colon cancer xenograft, for instance, all three proliferation markers were expressed in opposition to the differentiation marker KRT20 (Supplementary Fig. 16). This observation was subsequently confirmed at the protein level by a systematic study of Ki67 and KRT20 expression in serial sections from human colorectal cancer tissues, where Ki67 expression was often inversely associated with KRT20 (Supplementary Fig. 17). These observations suggest that, in many cases, bulk short-term tumor growth is principally driven by a specific subset of the cancer cell population, characterized by a gene-expression repertoire characteristic of more immature cell compartments. This concept has important implications for the modeling of tumor growth kinetics and the response to anti-tumor drugs in different experimental settings. Although very frequent, this feature is not necessarily absolute, as we have observed exceptions characterized either by homogenous expression of KRT20 in the almost entirety of the malignant epithelium or by complete absence of it (Supplementary Fig. 17, SU87 and SU98, respectively). In accordance with our model, however, tumors characterized by the complete absence of KRT20 expression were very poorly differentiated and contained high percentages of Ki67+ cells (Supplementary Fig. 17, SU98).
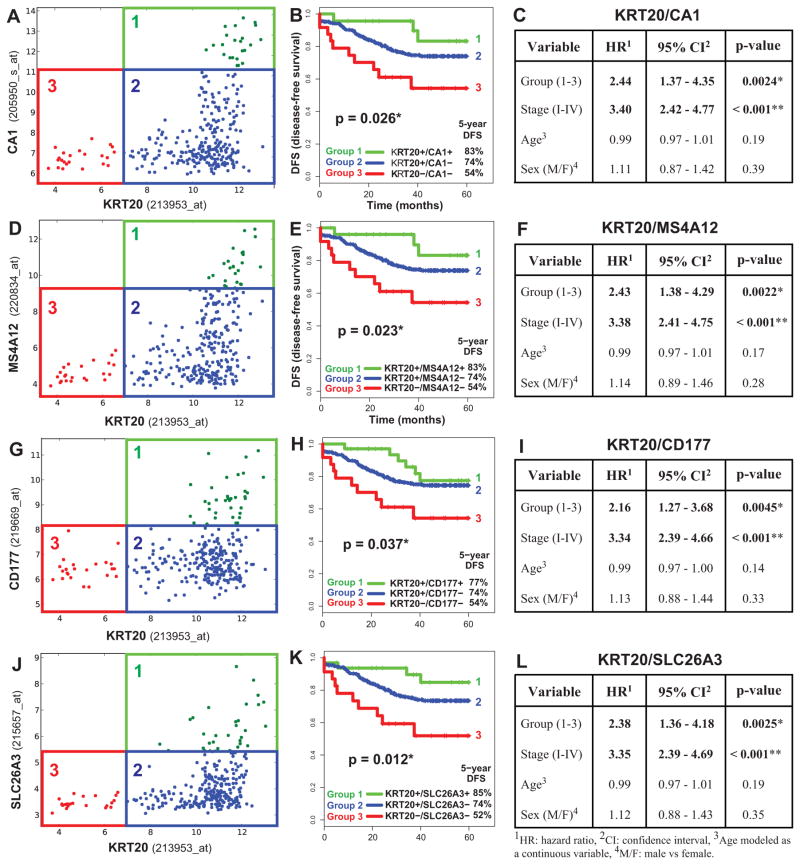
We next decided to test whether these novel insights in the functional anatomy of the colon epithelium could be used as a guide to develop clinically useful information. We evaluated whether quantitative expression levels of genes associated with differentiation processes could be used as a substitute measure for the cellular composition of the corresponding tumors and thereby serve to stratify colon cancer patients and predict prognosis. Our SINCE-PCR data identified a set of sensitive and exclusive markers of “top-of-the-crypt” CA1+/SLC26A3+ cells (i.e. CA1, MS4A12, CD177, SLC26A3). It also implicated KRT20 as a more promiscuous differentiation marker, whose expression is high in CA1+/SLC26A3+ cells and a subset of MUC2+/TFF3high cells, is absent in LGR5+/ASCL2+ cells, and is inversely associated to that of proliferation markers (MKI67, TOP2A, BIRC5). In addition, KRT20 expression can be easily detected by immunohistochemistry and is commonly used as a diagnostic marker in surgical pathology 38, thus representing an attractive candidate for further clinical applications 39.
Our first analysis on a pool of 1568 independent human colon gene-expression arrays revealed that expression levels for genes characteristic of the CA1+/SLC26A3+ cell population are strongly correlated (Supplementary Fig. 18), and that the relationship between the expression of these “top-of-the-crypt” genes and KRT20 is described by a structured distribution, where tumors expressing high levels of “top-of-the-crypt” genes (top-crypthigh) represent a subset of KRT20+ ones, and where tumors expressing low-to-negative levels of “top-of-the-crypt” genes (top-cryptneg/low) can be clearly separated into two groups: KRT20+ and KRT20neg (Supplementary Fig. 7). Interestingly, the KRT20neg colon cancer subset was also characterized by higher gene-expression levels of ALCAM/CD166 (Supplementary Fig 19), a surface marker highly expressed by cancer cell subsets endowed with high tumorigenic potential in mouse xenotransplantation models 37.
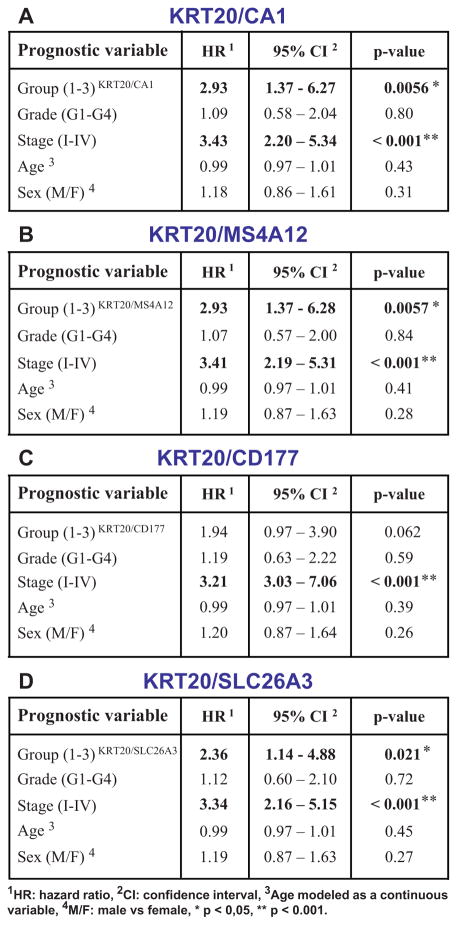
We then developed software (Hegemon, “hierarchical exploration of gene expression microarrays on-line”) to analyze the survival outcomes of human colon cancer patients after stratification into distinct gene-expression subsets, based on the expression of KRT20 and one of the marker genes of CA1+/SLC26A3+ “top-of-the-crypt” cells (Figure 4). These subsets, or gene-expression groups, were numbered from more to less mature (Group 1: KRT20+/top-crypthigh; Group 2: KRT20+/top-cryptneg/low; Group 3, KRT20neg/top-cryptneg/low). We used a computer-assisted method to determine the threshold level between positive and negative expression, based on the StepMiner algorithm (Supplementary Fig. 20) 40 and we compared the clinical outcome of colon cancer patients in the three groups, using a pool of three independent datasets, containing 299 patients of different clinical stages (AJCC Stage I–IV/Duke’s Stage A–D) from the H. Lee Moffit Cancer Center, the Vanderbilt Medical Center and the Royal Melbourne Hospital 41, 42, all of which annotated with disease-free survival (DFS) data. Interestingly, the analysis showed that the three patient groups identified by these simple two-gene classifiers displayed substantially different clinical outcomes, and that an increasingly immature gene-expression profile corresponded to a progressively worse prognosis (Fig. 4). This result was independent of the gene chosen as marker of CA1+/SLC26A3+ cells (i.e. CA1, MS4A12, CD177, SLC26A3) and, most importantly, a multivariate analysis indicated that the prognostic effect of the two-gene grouping system was not confounded by stage or other clinical variables (Fig. 4). Interestingly, tumors with a more immature gene-expression profile (Group 3, KRT20neg/top-cryptneg/low) were enriched in tumors with high pathological grade (G3-G4; Supplementary Fig. 21) and micro-satellite instability status (MSI; Supplementary Fig. 22). These enrichments, however, did not confound the prognostic effect of the two-gene classifier system, as the high hazard-ratios associated to more immature gene-expression groups remained statistically significant, and superior to those of higher pathological grade, when the two parameters were directly compared in multivariate analysis (Fig. 5), and because MSI+ tumors are known to be usually associated to a better, rather than worse, prognosis 43. Most interestingly, the prognostic effect of the two-gene classifier system was also independent of the recently described multi-gene EphB2 intestinal stem cell signature 19, and was associated with comparable, if not superior, hazard-ratios (Supplementary Fig. 23).
Figure 4. KRT20 and “top-crypt” genes can be used as novel prognostic markers in colorectal cancer patients.
To evaluate whether genes identified by SINCE-PCR as differentially expressed during normal colon differentiation can be used as prognostic markers for colon cancer patients, we analyzed a pooled database of 299 primary colon cancer gene-expression arrays annotated with disease-free survival (DFS) data (Jorissen and Smith, Supplementary Table 1). First, we used the Hegemon software to graph individual arrays according to the expression levels of KRT20 and one of four genes characteristic of “top-of-the-crypt” CA1+/SLC26A3+ enterocyte-like cells (A, CA1; D, MS4A12; G, CD177; J, SLC26A3) and we exploited the StepMiner algorithm to define gene-expression thresholds. In all four instances, three distinct gene-expression groups could be visualized: Group 1 (green), defined as KRT20+/CA1high, KRT20+/MS4A12 high, KRT20+/CD177+ or KRT20+/SLC26A3+, respectively; Group 2 (blue), defined as KRT20+/CA1neg/low, KRT20+/MS4A12neg/low, KRT20+/CD177neg or KRT20+/SLC26A3neg, respectively; Group 3 (red), defined as KRT20neg/CA1neg/low, KRT20neg/MS4A12neg/low, KRT20neg/CD177neg or KRT20neg/SLC26A3neg, respectively. In all instances, an increasingly immature gene-expression profile corresponded to a progressively worse prognosis (B, F, H, K). Multivariate analysis of survival data indicated that the prognostic effect of these “gene-expression groups” is not confounded by clinical stage, age or sex (C, F, I, L; * p < 0.05, ** p < 0.001).
Figure 5. The prognostic effect of “KRT20/top-crypt” gene-expression groups is not confounded by pathological grade and is associated to higher hazard ratios.
A direct comparison of the prognostic effect of gene-expression groups identified based on the “KRT20/top-crypt” two-gene scoring system with that of traditional pathological grading, using multivariate analysis based on the Cox proportional hazards model, was performed on a subset database of 181 microarrays annotated with grading information (Smith database, n=181, see Supplementary Table 1). The analysis indicated that the prognostic effect of “KRT20/top-crypt” gene-expression groups is not confounded by and is associated to higher hazard-ratios (HR) as compared to traditional pathological grade, independently of the gene chosen as marker of the “top-of-the-crypt” CA1+/SLC26A3+ enterocyte-type cell population, with the only exception of CD177 (A, CA1; B, MS4A12; C, CD177; D, SLC26A3; * p-value < 0.05, ** p-value < 0.001).
DISCUSSION
In this study, we implemented a novel approach to study the cellular composition of solid tissues, based on the high-throughput parallel analysis of the gene-expression repertoire of single-cells sorted by flow cytometry. We used this methodology to visualize the distinct cellular subsets of the human colon epithelium and to discover novel gene expression markers to define them. We then analyzed human colorectal tumors, both benign and malignant, and measured their perturbations in terms of cell lineage composition and maturation. We showed that tumor tissues contain multiple cell types whose transcriptional identities mirror those of the cellular lineages of the normal epithelium. Moreover, we showed that tumor tissues generated from a single cell can recapitulate the lineage diversity of parent tumors, demonstrating that multi-lineage differentiation represents a key source of in vivo functional and phenotypic cancer cell heterogeneity.
Using these concepts as a guide, we identified novel biological subsets of human colorectal cancer, based on their positive or negative expression of genes characteristic of specific cell types. Importantly, these novel biological subsets were associated to substantially different clinical outcomes, and could be identified by a simple two-gene classifier system. This novel prognostic scoring system appeared independent of and superior to traditional pathological grading, which is, to this date, among the few prognostic parameters used to design therapeutic algorithms for colon cancer patients 44. Due to its superior predictive value, as well as to its simplicity and quantitative nature, this two-gene scoring system has the potential to move beyond the realm of purely experimental medicine and become a viable candidate for clinical applications.
METHODS
Human primary tissues and colon cancer xenografts
Human primary colon tissues, both normal and malignant, were collected according to guidelines from Stanford University’s institutional review board. Human colon cancer xenograft lines were established and serially passaged in immunodeficient mice as previously described37. Human colon cancer tissues used in this study, either from primary samples or xenograft lines, are listed in Supplementary Table 4, together with clinical information related to corresponding patients. Solid tissues were disaggregated in single-cell suspensions and analyzed by flow cytometry following our previously published protocols37. A detailed description of the protocols used for the establishment and serial passage in mice of human colon cancer xenograft lines, and for the disaggregation and flow cytometry analysis of solid tissues, can be found in the Supplementary Methods.
Cell lines
Calibration experiments to measure accuracy and precision of single-cell sorting by flow cytometry, as well as to measure the single-cell sensitivity of the SINCE-PCR method, were performed on a clone of the HCT116 human colon cancer cell line infected with the pLentiLox3.7 lentivirus (pLL3.7, Addgene plasmid #11795, http://www.addgene.org), which encodes for the enhanced green fluorescent protein (EGFP). HCT116 cells are available from the American Tissue-type Culture Collection (ATCC; catalog number CCL-247, http://www.atcc.org). Human colon cancer cell lines were maintained in RPMI-1640 medium, supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, 120 μg/ml penicillin, 100 μg/ml streptomycin, 20 mM Hepes and 1mM Sodium Pyruvate, as previously described 45.
SINCE-PCR
Single cell gene-expression experiments were performed using Fluidigm’s M96 quantitative PCR (qPCR) DynamicArray™ microfluidic chips (Fluidigm, South San Francisco, CA). Single cells (n = 1) were sorted by FACS into individual wells of 96-well PCR plates using a protocol built-in within the FACSAriaII flow cytometer’s software package (BD Biosciences, San Jose, CA) with appropriate adjustments (device: 96-well plate; precision: single-cell; nozzle: 130 μm). Each 96-well plate was pre-loaded with 5μl/well of CellsDirect PCR mix and 0.1μl/well (2 U) of SuperaseIn RNase Inhibitor (Invitrogen, Carlsbad, CA). Following single-cell sorting, each well was supplemented with 1 μl of SuperScript III RT/Platinum Taq (Invitrogen), 1.5 μl of Tris-EDTA (TE) buffer and 2.5 μl of a mixture of 96 pooled TaqMan® assays (Applied Biosystems, Foster City, CA) containing each assay at 1:100 dilution. Single-cell mRNA was directly reverse transcribed into cDNA (50°C for 15 min., 95°C for 2 min.), pre-amplified for 20 PCR cycles (each cycle: 60°C for 4 min., 95°C for 15 sec) and finally diluted 1:3 with TE buffer. A 2.25 μl aliquot of amplified cDNA was then mixed with 2.5 μl of TaqMan qPCR mix (Applied Biosystems) and 0.25 μl of Fluidigm “sample loading agent”, then inserted into one of the chip “sample” inlets. Individual TaqMan® assays were diluted at 1:1 ratios with TE. A 2.5 μl aliquot of each diluted TaqMan® assay was mixed with 2.5 μl of Fluidigm “assay loading agent” and individually inserted into the chip “assay” inlets. Samples and probes were loaded into M96 chips using a HX IFC Controller (Fluidigm), then transferred to a Biomark real-time PCR reader (Fluidigm) following the manufacturer’s instructions. A list of the 57 TaqMan® assays used in this study can be found in Supplementary Table 2. A detailed description of both the SINCE-PCR protocol and the methodology used for the screening and selection of the 57 TaqMan® assays can be found in the Supplementary Methods.
Analysis and graphic display of SINCE-PCR data
SINCE-PCR data were analyzed and displayed using MATLAB® (MathWorks Inc., Natick, MA) as summarized in Supplementary Figure 2. A minimum of 336 cells were analyzed for each phenotypic population, corresponding to 4 PCR plates, each containing 84 single-cells (84 × 4 = 336), 8 positive and 4 negative controls. Results from cells not expressing ACTB (β-actin) and GAPDH (Glyceraldehyde 3-phosphate dehydrogenase), or expressing them at extremely low values (Ct >35), were removed from the analysis. Gene-expression results were normalized by mean centering and dividing by 3 times the standard deviation (3 SD) of expressing cells (Supplementary Fig. 2), and subsequently visualized using both hierarchical clustering and principal component analysis (PCA)12, 46. Hierarchical clustering was performed on both cells and genes, based on Euclidean or correlation distance metric and complete linkage. Positive or negative associations among pairs of genes were tested by Spearman correlation, and p-values calculated based on 10.000 permutations. Both hierarchical clustering and PCA were based on the results for 47 differentially expressed genes (51 assays), and excluded results from housekeeping genes (ACTB, GAPDH, EpCAM) and proliferation-related genes (MKI67, TOP2A, BIRC5/Survivin) to avoid noise based on proliferation status. A detailed description of the methods applied for analysis and graphic display of SINCE-PCR data, including the method to compare hierarchical clustering and PCA results, can be found in the Supplementary Methods
Immunohistochemistry and immunofluorescence
Paraffin-embedded tissue sections were stained with anti-human CK20 (clone Ks20.8, DakoCytomation), MUC2 (clone Ccp58, Fitzgerald Industries), Ki67 (clone MIB-1, DakoCytomation), CEACAM1/CD66a (clone 283340; R&D Systems) and SLC26A3 (lot #R32905, Sigma Life Science) antibodies, according to manufacturers’ instructions. Frozen tissue sections were stained with an anti-human CD177 antibody (clone MEM-166, BD Biosciences) followed by secondary staining with goat anti-mouse IgG-Alexa488 (Invitrogen). A detailed description of immunohistochemistry and immunofluorescence protocols can be found in the Supplementary Methods.
Generation and characterization of monoclonal tumors
EpCAMhigh/CD44+ human colon cancer cells were infected with the pLentiLox3.7 lentivirus (Addgene plasmid #11795, http://www.addgene.org)47. Monoclonal origin of tumors originated from single (n =1) lentivirus-infected EpCAMhigh/CD44+ cancer cells was confirmed by ligation-mediated PCR (LM-PCR)48, followed by DNA sequencing of LM-PCR amplification products. Tumorigenicity experiments were performed in NOD/SCID/IL2Rγ−/− immunodeficient mice in accordance with previously published protocols 37,49, 50, and Stanford University’s institutional animal welfare guidelines. A detailed description of the methods applied for lentivirus infection of cancer cells, LM-PCR characterization of lentivirus integration sites and in vivo tumorigenicity experiments can be found in the Supplementary Methods.
Mining of gene-expression arrays using Boolean implication analysis
Publicly available human gene-expression arrays were downloaded from NCBI’s GEO (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo), normalized and cross-checked for redundancies, as previously described 20. Gene-expression arrays included in this study are listed in Supplementary Table 1. Gene-expression thresholds between positive and negative samples were defined using the StepMiner algorithm 40, and Boolean implication relationships between pairs of genes using the BooleanNet software 20. Differences in gene-expression levels among different sample groups were evaluated using box-plots and tested for statistical significance using a 2-sample t-test (2-tailed). Correlations between two genes’ expression levels were measured using Pearson correlation coefficients. A detailed description of the procedures used for collection, normalization, purging and Boolean implication analysis of gene-expression arrays can be found in the Supplementary Methods.
Survival analysis and other statistical tests
Associations between gene-expression profiles and patient survival outcomes were investigated using a novel bioinformatic tool, named “Hegemon” for “hierarchical exploration of gene expression microarrays on-line”. Hegemon is an upgrade of the BooleanNet software, where individual gene-expression arrays, after being plotted on a two-axis chart based on the expression of two given genes 20, can be automatically grouped, stratified and compared for survival outcomes, using both Kaplan-Meier survival curves and multivariate analysis based on the Cox proportional hazards method. Differences in Kaplan-Meier curves were tested for statistical significance using the Log-rank test. Survival analysis was performed on a gene-expression database created by pooling information from two GEO datasets (GSE14333, GSE17538; Supplementary Table 1) 41, 42. This database contains disease-free survival (DFS) information on 299 patients from three independent institutions: H. Lee Moffit Cancer Center (n = 164), Vanderbilt Medical Center (n = 55) and Royal Melbourne Hospital (n = 80). Enrichment of selected pathological or molecular features, such as high pathological grade (G3–G4) or microsatellite instability (MSI), in groups characterized by immature gene-expression patterns (e.g. Group 3, KRT20neg/top-cryptneg/low) was measured using odds-ratios (OR) and tested for significance using Pearson’s χ2 test. A detailed description of the procedures used for patient stratification in gene-expression groups, comparison of survival outcomes and evaluation of enrichment of specific features in tumors belonging to a specific gene-expression group can be found in the Supplementary Methods.
Supplementary Material
1
Acknowledgments
This study was supported by NIH grants U54-CA126524 and P01-CA139490 (to S.R.Q. and M.F.C.) and the NIH Director’s Pioneer Awards (to S.R.Q.). P.D. was supported by a training grant from the California Institute for Regenerative Medicine (CIRM) and by a BD Biosciences Stem Cell Research Grant (Summer 2011). T.K. was supported by a fellowship from the Machiah Foundation. D.S. was supported by NIH grant K99-CA151673, by DoD grant W81XWH-10-1-0500 and a grant from the Siebel Stem Cell Institute and the Thomas and Stacey Siebel Foundation. We wish to thank Robert Tibshirani and Daniela Witten for helpful suggestions about data analysis. We are grateful to Luigi Warren, Richard A. White IIIrd, Edward Gilbert, Patricia Lovelace, Marissa Palmor, Coralie Donkers and Stephen P. Miranda for helpful discussion and technical support in many moments during the completion of this study.
Footnotes
AUTHOR CONTRIBUTIONS
P.D., T.K., D.S., M.F.C. and S.R.Q. conceived the study and designed the experiments. P.S.R., M.E.R., A.A.L., M.Z., N.F.N, M. v. d. W. and H.C. provided intellectual guidance in the design of selected experiments. P.D., T.K., D.S., P.S.R., A.A.L., S.S., J.O., D.M.J., D.Q., J.W., Y.S. and S.H. performed the experiments. P.D., T.K., D.S., N.F.N., Y.S., M.F.C. and S.R.Q analyzed the data and/or provided intellectual guidance in their interpretation. J.B., A.A.S. and B.V. provided samples and reagents. P.D., T.K., D.S., M.F.C. and S.R.Q. wrote the paper.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 3.Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 4.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 5.Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6:2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 6.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochimica et biophysica acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirkland SC. Clonal origin of columnar, mucous, and endocrine cell lineages in human colorectal epithelium. Cancer. 1988;61:1359–1363. doi: 10.1002/1097-0142(19880401)61:7<1359::aid-cncr2820610714>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 8.Odoux C, et al. A stochastic model for cancer stem cell origin in metastatic colon cancer. Cancer Res. 2008;68:6932–6941. doi: 10.1158/0008-5472.CAN-07-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vermeulen L, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci U S A. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Developmental cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 13.White AK, et al. High-throughput microfluidic single-cell RT-qPCR. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiao YF, Nakamura S, Sugai T, Yamada N, Habano W. Serrated adenoma of the colorectum undergoes a proliferation versus differentiation process: new conceptual interpretation of morphogenesis. Oncology. 2008;74:127–134. doi: 10.1159/000151359. [DOI] [PubMed] [Google Scholar]
- 15.Wielenga VJ, et al. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol. 1999;154:515–523. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prall F, et al. CD66a (BGP), an adhesion molecule of the carcinoembryonic antigen family, is expressed in epithelium, endothelium, and myeloid cells in a wide range of normal human tissues. J Histochem Cytochem. 1996;44:35–41. doi: 10.1177/44.1.8543780. [DOI] [PubMed] [Google Scholar]
- 17.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 18.Becker L, Huang Q, Mashimo H. Immunostaining of Lgr5, an intestinal stem cell marker, in normal and premalignant human gastrointestinal tissue. TheScientificWorldJournal. 2008;8:1168–1176. doi: 10.1100/tsw.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merlos-Suarez A, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell stem cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 20.Sahoo D, Dill DL, Gentles AJ, Tibshirani R, Plevritis SK. Boolean implication networks derived from large scale, whole genome microarray datasets. Genome biology. 2008;9:R157. doi: 10.1186/gb-2008-9-10-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoglund P, et al. Mutations of the Down-regulated in adenoma (DRA) gene cause congenital chloride diarrhoea. Nat Genet. 1996;14:316–319. doi: 10.1038/ng1196-316. [DOI] [PubMed] [Google Scholar]
- 22.Fischer H, Stenling R, Rubio C, Lindblom A. Differential expression of aquaporin 8 in human colonic epithelial cells and colorectal tumors. BMC physiology. 2001;1:1. doi: 10.1186/1472-6793-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koslowski M, Sahin U, Dhaene K, Huber C, Tureci O. MS4A12 is a colon-selective store-operated calcium channel promoting malignant cell processes. Cancer Res. 2008;68:3458–3466. doi: 10.1158/0008-5472.CAN-07-5768. [DOI] [PubMed] [Google Scholar]
- 24.Noah TK, Kazanjian A, Whitsett J, Shroyer NF. SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Experimental cell research. 2010;316:452–465. doi: 10.1016/j.yexcr.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gregorieff A, et al. The ets-domain transcription factor Spdef promotes maturation of goblet and paneth cells in the intestinal epithelium. Gastroenterology. 2009;137:1333–1345. e1331–1333. doi: 10.1053/j.gastro.2009.06.044. [DOI] [PubMed] [Google Scholar]
- 26.van der Flier LG, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 27.Ezhkova E, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park IK, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 29.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell stem cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beider K, Abraham M, Peled A. Chemokines and chemokine receptors in stem cell circulation. Front Biosci. 2008;13:6820–6833. doi: 10.2741/3190. [DOI] [PubMed] [Google Scholar]
- 32.Jensen KB, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell stem cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalla-Favera R, Wong-Staal F, Gallo RC. Onc gene amplification in promyelocytic leukaemia cell line HL-60 and primary leukaemic cells of the same patient. Nature. 1982;299:61–63. doi: 10.1038/299061a0. [DOI] [PubMed] [Google Scholar]
- 34.Hoey T, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell stem cell. 2009;5:168–177. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 35.Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Losi L, Baisse B, Bouzourene H, Benhattar J. Evolution of intratumoral genetic heterogeneity during colorectal cancer progression. Carcinogenesis. 2005;26:916–922. doi: 10.1093/carcin/bgi044. [DOI] [PubMed] [Google Scholar]
- 37.Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oien KA. Pathologic evaluation of unknown primary cancer. Seminars in oncology. 2009;36:8–37. doi: 10.1053/j.seminoncol.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 39.Lugli A, Tzankov A, Zlobec I, Terracciano LM. Differential diagnostic and functional role of the multi-marker phenotype CDX2/CK20/CK7 in colorectal cancer stratified by mismatch repair status. Mod Pathol. 2008;21:1403–1412. doi: 10.1038/modpathol.2008.117. [DOI] [PubMed] [Google Scholar]
- 40.Sahoo D, Dill DL, Tibshirani R, Plevritis SK. Extracting binary signals from microarray time-course data. Nucleic Acids Res. 2007;35:3705–3712. doi: 10.1093/nar/gkm284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorissen RN, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith JJ, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46:2788–2798. doi: 10.1016/j.ejca.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Bardia A, et al. Adjuvant chemotherapy for resected stage II and III colon cancer: comparison of two widely used prognostic calculators. Seminars in oncology. 2010;37:39–46. doi: 10.1053/j.seminoncol.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalerba P, et al. Reconstitution of human telomerase reverse transcriptase expression rescues colorectal carcinoma cells from in vitro senescence: evidence against immortality as a constitutive trait of tumor cells. Cancer Res. 2005;65:2321–2329. doi: 10.1158/0008-5472.CAN-04-3678. [DOI] [PubMed] [Google Scholar]
- 46.Ringner M. What is principal component analysis? Nat Biotechnol. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 47.O’Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. Journal of virology. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang GP, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishizawa K, et al. Tumor-Initiating Cells Are Rare in Many Human Tumors. Cell stem cell. 2010;7:279–282. doi: 10.1016/j.stem.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quintana E, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1