A Quantitative Spatial Proteomics Analysis of Proteome Turnover in Human Cells (original) (raw)
Abstract
Measuring the properties of endogenous cell proteins, such as expression level, subcellular localization, and turnover rates, on a whole proteome level remains a major challenge in the postgenome era. Quantitative methods for measuring mRNA expression do not reliably predict corresponding protein levels and provide little or no information on other protein properties. Here we describe a combined pulse-labeling, spatial proteomics and data analysis strategy to characterize the expression, localization, synthesis, degradation, and turnover rates of endogenously expressed, untagged human proteins in different subcellular compartments. Using quantitative mass spectrometry and stable isotope labeling with amino acids in cell culture, a total of 80,098 peptides from 8,041 HeLa proteins were quantified, and their spatial distribution between the cytoplasm, nucleus and nucleolus determined and visualized using specialized software tools developed in PepTracker. Using information from ion intensities and rates of change in isotope ratios, protein abundance levels and protein synthesis, degradation and turnover rates were calculated for the whole cell and for the respective cytoplasmic, nuclear, and nucleolar compartments. Expression levels of endogenous HeLa proteins varied by up to seven orders of magnitude. The average turnover rate for HeLa proteins was ∼20 h. Turnover rate did not correlate with either molecular weight or net charge, but did correlate with abundance, with highly abundant proteins showing longer than average half-lives. Fast turnover proteins had overall a higher frequency of PEST motifs than slow turnover proteins but no general correlation was observed between amino or carboxyl terminal amino acid identities and turnover rates. A subset of proteins was identified that exist in pools with different turnover rates depending on their subcellular localization. This strongly correlated with subunits of large, multiprotein complexes, suggesting a general mechanism whereby their assembly is controlled in a different subcellular location to their main site of function.
Cells can regulate proteins via phosphorylation and other reversible modifications, and through altering protein level by changing the rate of synthesis and/or degradation (1). DNA microarrays are used extensively for analysis of gene expression at the RNA level. Although abundant mRNAs usually result in high protein levels (2), the general correlation between mRNA levels and protein abundance is often poor (3). The regulatory complexity of mRNA translation and protein stability emphasizes the need for direct measurements of protein levels. Mass spectrometry-based proteomics has emerged as the technology of choice for studying proteins directly, allowing not only identification of proteins and post-translational modifications, but also quantitative comparisons of how relative protein levels change in cells under different conditions (4).
There are two main pathways for intracellular protein degradation, i.e. the proteasome and autophagy-lysosomal systems. The ubiquitin-proteasome pathway identifies proteins for degradation by attachment of poly-ubiquitin tags, which targets the modified proteins for degradation by the proteasome (5). In the autophagy-lysosomal system proteins destined for degradation are captured within membrane bound organelles (phagosomes) for bulk digestion (1). Cell growth requires a net increase in total protein and thus higher levels of translation than degradation. Maintaining protein levels at steady-state also involves continuous protein synthesis, balanced with degradation. Protein turnover rates can range from under 10 mins to over a hundred hours (1).
Some biological processes involve constant cycles of protein production and rapid degradation. For example, despite continuous synthesis of the tumor suppressor p53, its constant rapid degradation results in low steady state levels under normal cell growth conditions (6). Upon oncogene activation, degradation of p53 is prevented through sequestration of the E3 ligase mdm2, causing a rapid increase in p53 levels independent of transcriptional activation. Control of protein degradation thus provides a flexible mechanism for the rapid activation of gene expression in mammalian cells.
The turnover rates of specific proteins can vary between different subcellular compartments. Using a combination of pulsed stable isotope labeling with amino acids in cell culture (SILAC)1 and fluorescence microscopy, it was shown that HeLa cells constantly import and degrade high levels of free ribosomal proteins in the nucleus. Ribosomal protein stability is dramatically increased upon assembly into ribosome subunits and export to the cytoplasm (7). Importantly, this shows that protein half-life values based only on analyses of whole cell extracts provide average values that can mask the existence of pools of protein with different properties.
Early studies of protein turnover relied on detecting incorporation of radiolabeled amino acids into newly translated proteins and either analyzed bulk protein turnover, or else turnover of individual proteins (8). Typically, proteins were labeled with [35S] methionine and pulse-chase experiments used to determine their rate of degradation after blocking protein synthesis. The use of protein synthesis inhibitors such as cycloheximide raises concerns whether the normal degradation processes, or other aspects of cellular activity, may also be disrupted. Mass spectrometry-based proteomics now allows determination of the turnover rates of large numbers of proteins in single experiments using pulse labeling with amino acids incorporating stable isotopes (7, 9–12).
SILAC is the use of stable isotopic atoms for quantitative mass spectrometry analysis (13, 14). This allows quantitative analyses of proteins by comparison of the mass of light and heavier forms of the same peptide containing amino acids with stable isotopes such as 13C, 2H, and 15N. These isotope-tagged amino acids are incorporated into proteins in vivo where typically arginine and lysine are replaced with corresponding heavy isotope-substituted forms (15). Cleavage at the substituted arginine or lysine by trypsin generates a peptide with a shift in mass relative to the control, “light” peptide and this is resolved and quantitated by mass spectrometry. The intensity ratio of “light” and “heavy” peptides correlate with the relative amount of the cognate protein from each sample. SILAC has been successful for quantitative analysis of cell and organelle proteomes and for comparative studies of protein modifications, and interactions (4) and to identify proteins isolated from mitotic chromosomes (16). We have used SILAC in combination with cell fractionation to generate “isotope-encoded” subcellular compartments allowing subcellular protein localization to be evaluated on a system-wide level (17). This spatial proteomics approach provides a high-throughput assay for the unbiased analysis of changes in subcellular protein localization arising in response to perturbations such as DNA damage and for comparing protein localization and responses in cell lines with different genotypes (18).
Here we combine an enhanced pulse SILAC approach with spatial proteomics to perform a system-wide analysis of protein turnover in cultured human cells. Protein abundance and the rates of protein synthesis, degradation and turnover have been measured in parallel for whole cells and for separate cytoplasmic, nuclear and nucleolar compartments, providing a cell-based functional annotation of the human proteome.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa cells were cultured as adherent cells in DMEM (Dulbeccos's modified eagle medium, Invitrogen, custom order) depleted of arginine and lysine. The DMEM was supplemented with 10% fetal bovine serum dialyzed with a cut-off of 10 kDa (Invitrogen, 26400–044), 100 U/ml penicillin/streptomycin, 2 mm l-Glutamine. Arginine and lysine was added in either light (Arg0, Sigma, A5006; Lys0, Sigma, L5501), medium (Arg6, Cambridge Isotope Lab (CIL), Andover, MA; CNM-2265; Lys4, CIL, DLM-2640), or heavy (Arg10, CIL, CNLM-539; Lys8, CIL, CNLM-291) form to a final concentration of 28 μg/ml for arginine and 49 μg/ml for lysine. Proteins were tested for >99% incorporation of the label after six passages by mass spectrometry (data not shown).
Cell Fractionation
Cytoplasm, nuclei, and nucleoli were prepared from HeLa cells essentially as previously described (19). Briefly, cells were washed three times with phosphate-buffered saline (PBS), resuspended in 5 ml buffer A (10 mm HEPES-KOH [pH 7.9], 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT), and dounce homogenized ten times using a tight pestle. Dounced nuclei were centrifuged at 228 × g for 5 min at 4 °C. The supernatant represents the cytoplasmic fraction. The nuclear pellet was resuspended in 3 ml 0.25 m sucrose, 10 mm MgCl2, and layered over 3 ml 0.35 m sucrose, 0.5 mm MgCl2, and centrifuged at 1430 × g for 5 min at 4 °C. The clean, pelleted nuclei were resuspended in 3 ml 0.35 m sucrose, 0.5 mm MgCl2, and sonicated for 6 × 10 s using a microtip probe and a Misonix XL 2020 sonicator at power setting 5. The sonication was checked using phase contrast microscopy, ensuring that there were no intact cells and that the nucleoli were readily observed as dense, refractile bodies. The sonicated sample was then layered over 3 ml 0.88 m sucrose, 0.5 mm MgCl2 and centrifuged at 2800 × g for 10 min at 4 °C. The pellet contained the nucleoli, while the supernatant consisted of the nucleoplasmic fraction. The nucleoli were then washed by resuspension in 500 μl of 0.35 m sucrose, 0.5 mm MgCl2, followed by centrifugation at 2000 × g for 2 min at 4 °C. Proteins were quantified using the Quant-IT protein assay (Invitrogen) and measured using a Qubit (Invitrogen).
Gel Electrophoresis and In-Gel Digestion
For each time point, proteins were reduced in 10 mm dithiothreitol (DTT) and alkylated in 50 mm iodoacetamide prior to boiling in loading buffer, and then separated by one-dimensional SDS-PAGE (4–12% Bis-Tris Novex mini-gel, Invitrogen) and visualized by colloidal Coomassie staining (Novex, Invitrogen). The entire protein gel lanes were excised and cut into 16 slices each. Every gel slice was subjected to in-gel digestion with trypsin (20). The resulting tryptic peptides were extracted by 1% formic acid, then 100% acetonitrile, lyophilized in a speedvac, and resuspended in 1% formic acid.
Liquid Chromatography-Tandem MS (LC-MS/MS)
Trypsin digested peptides were separated using an Ultimate U3000 (Dionex Corporation) nanoflow LC-system consisting of a solvent degasser, micro and nanoflow pumps, flow control module, UV detector, and a thermostated autosampler. Ten microliters of sample (a total of 2 μg peptide) was loaded with a constant flow of 20 μl/min onto a PepMap C18 trap column (0.3 mm id × 5 mm, Dionex Corporation). After trap enrichment peptides were eluted onto a PepMap C18 nano column (75 μm × 15 cm, Dionex Corporation) with a linear gradient of 5–35% solvent B (90% acetonitrile with 0.1% formic acid) over 65 min with a constant flow of 300 nl/min. The performance liquid chromatography (HPLC) system was coupled either to a LTQ OrbiTrap XL (Thermo Fisher Scientific Inc), or to a LTQ OrbiTrap Velos, via a nano ES ion source (Proxeon Biosystems). The spray voltage was set to 1.2 kV and the temperature of the heated capillary was set to 200 °C. Full scan MS survey spectra (m/z 335–1800) in profile mode were acquired in the Orbitrap with a resolution of 60,000 after accumulation of 500,000 ions. The five most intense peptide ions from the preview scan in the Orbitrap were fragmented by collision induced dissociation (normalized collision energy 35%, activation Q 0.250 and activation time 30 ms) in the LTQ after the accumulation of 10,000 ions. Maximal filling times were 1000 ms for the full scans and 150 ms for the MS/MS scans. Precursor ion charge state screening was enabled and all unassigned charge states as well as singly charged species were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 90 s and a relative mass window of 10 ppm. The lock mass option was enabled for survey scans to improve mass accuracy (21). Data were acquired using the XCalibur software.
Quantification and Bioinformatic Analysis
Quantitation was performed using the program MaxQuant version 1.1.1.14 (22, 23). The derived peak list generated by Quant.exe (the first part of MaxQuant) was searched using Andromeda as the database search engine for peptide identifications against the International Protein Index (IPI) human protein database version 3.68 containing 89,422 proteins, to which 175 commonly observed contaminants and all the reversed sequences had been added. The initial mass tolerance was set to 7 p.p.m. and MS/MS mass tolerance was 0.5 Da. Enzyme was set to trypsin/p with 2 missed cleavages. Carbamidomethylation of cysteine was searched as a fixed modification, whereas _N_-acetyl protein and oxidation of methionine were searched as variable modifications. Identification was set to a false discovery rate of 1%. To achieve reliable identifications, all proteins were accepted based on the criteria that the number of forward hits in the database was at least 100-fold higher than the number of reverse database hits, thus resulting in a false discovery rate of less than 1%. A minimum of 2 peptides were quantified for each protein. Protein isoforms and proteins that cannot be distinguished based on the peptides identified are grouped and displayed on a single line with multiple IPI numbers (see supplementary tables).
PepTracker Spatial Viewer Database
The PepTracker turnover and spatial viewer consists of a web-based, multi-tier architecture, where the data storage, server-side logic, and user interface are separate components (see; http://www.peptracker.com/). The data storage is implemented as a fully relational Oracle database (Oracle Database 10g Enterprise Edition Release 10.2.0.5.0). This database holds turnover details, both at the protein and peptide level. The server-side logic and client interface reside on an Apache web server (Version 2.2.3 - CentOS Linux Distribution). The server-side logic is implemented using Python (Version 2.6.4), structured using the Django framework (Version 1.2.1, http://www.djangoproject.com/). The Django framework enforces code to follow the model view controller design pattern, thus the functionality within the application is separated from the overall look and feel of the application, ensuring a more customizable solution. The server-side logic makes use of complex Structured Query Language (SQL) in order to communicate with the database and extract the relevant data required by the user interface. It does so using SQLAlchemy (http://www.sqlalchemy.org/), a Python Structured Query Language toolkit and object relational mapper. The Django framework provides the ability to setup html templates that form the user interface. These templates are customized on the fly using the data passed to them from the server-side logic. The templates are coded in HyperText Markup Language (HTML), with Javascript additions to enhance functionality. Specifically, the viewer makes use of the JQuery library (http://jquery.com/) and Google Visualization API (http://code.google.com/apis/visualization/interactive_charts.html) to provide additional elements, such as interactive charts. The interface also includes an Adobe Flex component that provides an interactive chart and cell map for users to navigate through the data. To further enhance the user experience the user interface performs dynamic requests to the server using REST (Representational State Transfer). These requests prevent HyperText Markup Languages pages from having to be completely redrawn and instead only the relevant sections of a page are updated. A detailed description of the PepTracker software will be published separately. Information on the data viewers, Protein Frequency Library and other PepTracker resources available currently and in future is provided at the website, www.peptracker.com.
RESULTS
Experimental Design
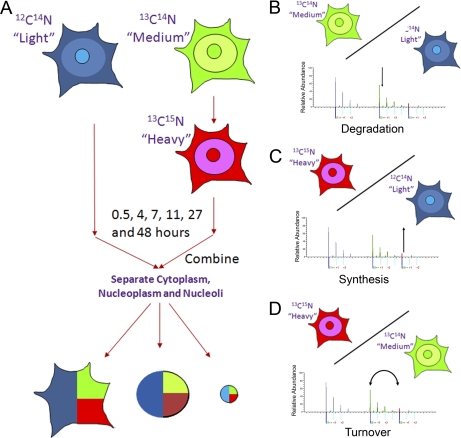
HeLa cells were grown in media containing arginine and lysine, either with the normal light isotopes of carbon, hydrogen and nitrogen (i.e. 12C14N) (light – “L”), or else with l-arginine-13C614N4 and l-lysine-2H4 (medium – “M”) for at least five cell divisions, resulting in >99% incorporation of the M amino acids (Fig. 1A). The culture media with the M amino acids is then replaced with media containing l-arginine- 13C6-15N4 and l-lysine-13C6-15N2 (heavy –“H”). H amino acids are pulsed into cells with M-labeled proteins for varying times, from 30 min to 48 h. For each peptide at each time point the fraction of H amino acids incorporated replacing the pre-existing M amino acids is determined by MS.
Fig. 1.
Pulse SILAC method. A, HeLa cells are cultured in different SILAC media containing either “light” (L), or “medium” (M) arginines and lysines until full incorporation of the amino acids. The medium of the cells growing with the “medium” amino acids is then changed for a “heavy” (H) medium. Cells are then harvested at different times, along with the equivalent cells growing in the “light” medium. Equal amounts of cells are then combined and separate cytoplasmic, nuclear, and nucleolar fractions were isolated from each time point. The resulting ratios: M/L isotopes over time measures the rate of protein degradation B, increase in the ratio: of H/L measures new protein synthesis C, and the change in the H/M ratio measures the rate of net protein turnover D.
Cells were harvested at 0.5, 4, 7, 11, 27, and 48 h time points following the H amino acid-pulse. At each time point, the pulsed cell sample was mixed with an equal number of HeLa cells grown in normal (i.e. light – “L”) culture media. This provides an internal control, allows separate measurement of protein synthesis, degradation, and turnover rates and facilitates normalization of the isotope incorporation data, thereby improving the accuracy of the measurements (supplementary Information). Moreover, this light sample enables the use of peptide ion intensity to estimate protein abundance, both in the whole cell, and in each subcellular compartment. The decreasing ratio of M/L isotopes over time measures the rate of protein degradation (Fig. 1B), whereas the increasing ratio of H/L measures protein translation (Fig. 1C) and the change in the H/M ratio measures the rate of net protein turnover (Fig. 1D). The 50% turnover time for each protein was also determined separately by analysis of the crossover between the respective synthesis and degradation curves and these values compared with the net turnover values obtained by measuring rates of change in H/M ratio.
The mixture of 50% L cells with 50% H/M cells was fractionated as described in the Methods section to generate separate cytoplasmic, nuclear and nucleolar fractions for each time point (Fig. 1A). All samples were solubilized with loading buffer, proteins separated using SDS-PAGE and the resulting gels cut into 16 equal pieces, trypsin digested, and analyzed by LC-MS/MS. Every sample was analyzed twice by mass spectrometry and the resulting ratios between light, medium, and heavy isotopic forms for each peptide were quantified using MaxQuant (22).
Protein Identification, Abundance, and Subcellular Localization
This analysis has identified and quantitated 80,098 peptides, mapped onto 8,041 endogenous HeLa cell proteins, yielding an average of ∼10 peptides per protein (see supplemental Table S1). The abundance of each protein was estimated based on the averaged peptide ion intensities from the control, light sample at each time point (24). Peptide intensity profiles were normalized from the top three peptides, based on their mean profile intensity (see supplementary Information).
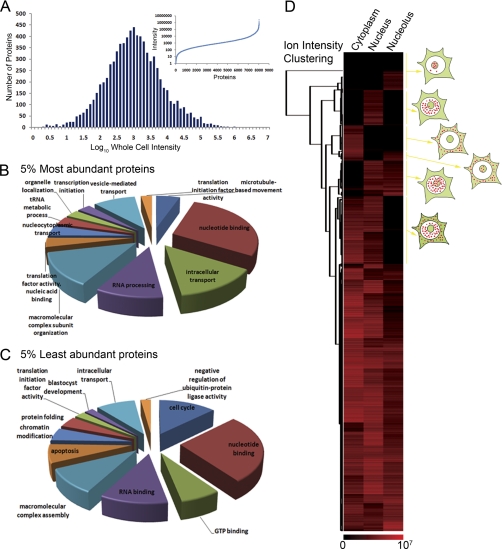
The protein abundance data span a dynamic range of intensity of ∼1 × 107 following a normal distribution with a mean intensity of ∼7000 (Fig. 2A). This shows the very large variation in copy number of endogenous human proteins expressed from different genes. Known abundant proteins, including nucleophosmin, histones, ribosomal proteins, actin, tubulin, GAPDH, and heat shock proteins, were among the top 1% highest intensity proteins. Histones are predominantly stably incorporated into nucleosomes with on average ∼150 million nucleosomes per human cell. Therefore, as histones showed ion intensities ∼1,000,000, we estimate that proteins with the lowest intensity values have a copy number ∼50–150 molecules per cell whereas the bulk of HeLa proteins are expressed at ∼1000–10,000 copies per cell (Fig. 2A). However, as these estimates are derived from averaging values over the cell population, there could be significant variation in the levels of proteins present at the single cell level.
Fig. 2.
Protein identification, abundance and subcellular localization. Peptide intensity profiles normalized from the top three peptides based on their mean profile intensity were used to measure protein abundance. A, A distribution plot with the protein count on the y axis and bins of 0.1 of the log10 intensity values on the x axis. The inset shows the distribution from the lowest intensity to the highest intensity protein with the intensity on the y axis and the protein number on the x axis. B, A gene ontology annotation analysis of the 5% most abundant proteins identified using functional clustering of biological processes and molecular functions (GO_BP and GO_MF). C, A gene ontology annotation analysis of the 5% lowest abundant proteins identified using functional clustering of biological processes and molecular functions. D, A hierarchical clustering was performed using the log10 value for intensity for the cytoplasm, the nucleus and the nucleolus and represented as a heat map. In each case high values are shown in red and low ratios in black.
Gene ontology molecular function annotation analysis of the 5% most abundant proteins identified factors involved in nucleotide binding, intracellular transport, RNA processing, and macromolecular complex subunit organization (Fig. 2B). Analysis of the 5% lowest abundance proteins revealed functions related to nucleotide binding, GTP binding, RNA binding, and cell cycle regulation (Fig. 2C). Although both the highest and lowest abundance protein groups had “nucleotide binding” as the largest molecular function class, the types of nucleotide binding proteins were different in each case. Thus, many transcription factors were included among the very low abundance proteins whereas histones and hnRNPs were prominent among the high abundance proteins. Over 40% of the lowest abundance proteins are either uncharacterized open reading frames (ORFs), or else proteins named only based on their molecular weight, or on a recognizable domain. In contrast, less than 1% of the most highly abundant proteins are uncharacterized ORFs or of unknown function.
The light peptide ion intensities measured in the fractionated subcellular compartments allow separate estimation of protein abundance in the cytoplasm, nucleus and nucleolus, providing a quantitative map of protein localization within the cell. A hierarchical clustering was performed and visualized as a heat map, using log10 intensity value for each compartment (Fig. 2D). For more than half of the proteins, intensity values were detected in more than one compartment. Relatively few proteins show equal distributions between two or three compartments (Fig. 2D). Thus, at steady state, most HeLa proteins are predominantly partitioned into specific subcellular locations. However, this does not exclude that proteins can shuttle between their major site of accumulation and other compartments.
Determination of Protein Turnover
We have used two methods to evaluate the time point at which 50% turnover has occurred for each protein (supplementary Information). The first method, relying on changes in the H/M isotope ratio, directly measures when 50% of the intensity signal for a peptide is M and 50% H, isotope. The corresponding protein turnover is defined here as the mean time for 50% incorporation of H amino acids for all peptides identified from that protein. The second method relies upon measuring the separate curves of synthesis and degradation rates for each protein, based on rates of change in H/L and M/L isotope ratios, and then identifying the point at which these curves cross, which corresponds to the time it takes for 50% of the protein to turn over (supplemental Fig. S2).
An interesting observation from the crossover method is that we measured an offset value (B) of ∼20%. This B value (see supplementary Information) reflects the fraction of M amino acids remaining in proteins once a steady state level of H amino acid incorporation is established. If H amino acid incorporation completely replaced the pre-existing M amino acids then the B value should be zero. The fact that it remains at ∼20% indicates persistent incorporation of M isotope-containing amino acids into proteins even after prolonged growth in H amino acid medium. This could result from recycling of M amino acids following degradation of the M-labeled proteins present at the start of the pulse and/or from an enduring intracellular M amino acid pool. Indeed, previous work has reported amino acid recycling from degraded proteins (25). To test whether the amino acid pool settles at ∼80% H amino acids, we analyzed the mass isotopomer distribution of peptides with missed trypsin cleavage to determine the level of peptides that contained both M + H amino acids (supplemental Fig. S7). This showed that ∼10–20% of peptides with missed cleavages consistently had both M and H amino acids in the same peptide, consistent with a precursor pool of ∼80–90% H amino acids (supplemental Fig. S7). Moreover, we found for these same missed cleavage peptides that there was virtually no combined M + H amino acids in the same peptide in an experiment where the SILAC medium was changed every hour over the time course (supplemental Fig. S7). We infer that without more than one replacement of the cell growth medium during the course of the experiment, the internal amino acid pool is likely not fully replaced with the externally supplied H amino acids.
A simple mathematical model of protein synthesis and degradation developed here (see supplementary Information for details) demonstrates that recycling of degraded proteins can lead to a nonzero offset in degradation curves (supplemental Fig. S3 and S4). To test this hypothesis, we analyzed whether the offset value B would decrease toward zero if during the time course of the pulse the external media containing H amino acids was repeatedly replaced (supplementary Information). This showed that replacing the media containing the H amino acids several times during the time course of the pulse resulted in the offset B reducing to ∼ 0, as expected for complete replacement of M with H amino acids (supplemental Figs. S1 and S6). We conclude that the intracellular pool of M amino acids either is not fully depleted when the medium is initially replaced, or else is replenished through recycling of amino acids from degradation of pre-existing M-labeled proteins, or both.
Another parameter that was determined was the respective protein half-life values, which represents here the time taken for 50% of the pool of each pre-existing protein species to be degraded (supplemental Table S2). We note that this study does not provide half-life values at the single molecule level but rather reflects average values for populations of protein molecules. The half-life values should reflect rates of protein degradation. To take into account the inevitable dilution effect on pools of pre-existing M-labeled proteins as a result of cell growth and new protein synthesis, rather than degradation, the half-lives were calculated using a formula that incorporates the growth rate measured here for HeLa cells growing in SILAC medium (supplementary Information). A comparison of the separate protein turnover and half-life values determined in this study showed that they are closely correlated (Pearson Correlation Coefficient 0.54). As the 50% turnover values more directly reflect both protein synthesis and degradation rates, and can be measured more accurately, we have focused our subsequent analyses specifically on a comparison of turnover values with other protein properties.
Distribution of Protein Turnover
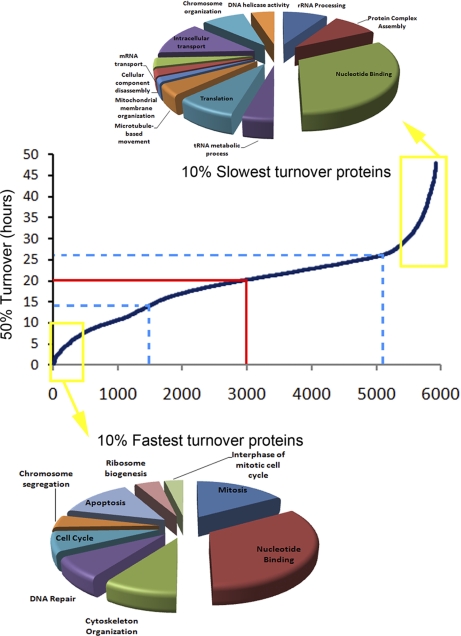
To compare proteins according to their 50% turnover value, proteins are shown from fastest to slowest 50% turnover value, represented as a scatter plot with the protein turnover on the y axis (Fig. 3). Approximately 60% of HeLa proteins have 50% turnover values clustered within 5 h of the average turnover rate of ∼20 h (Fig. 3, blue lines). However, if we correct for protein abundance, it takes ∼24 h for 50% turnover of the total HeLa proteome, because a subset of abundant proteins has turnover values longer than the mean of ∼20 h. This is close to the cell doubling time under the growth conditions used which we measured to be 24.67 h for HeLa cells growing in the SILAC medium, consistent with approximate doubling of the protein content as the cell divides (Fig. 3, red line).
Fig. 3.
Distribution of protein turnover. Proteins were sorted on the x axis from fastest to slowest turnover and represented as a scatter plot with the 50% protein turnover value on the y axis. Approximately 60% (blue lines) of the HeLa proteins show a 50% turnover rate within 5 h of the average of ∼20 h (red lines). Functional annotation clustering of gene ontology terms for the 10% proteins with the fastest (bottom) and slowest (top) turnover rates are shown as pie charts, using the number of proteins as weight for each annotation.
Functional annotation clustering of gene ontology terms for the fastest and slowest turnover rates showed specific enrichments of proteins with similar functions or characteristics (26, 27) (Fig. 3). The slowest turnover proteins have a wide range of functions. However, most are either present in large, abundant and stable protein complexes, such as ribosome and spliceosome subunits, RNA polymerase II, the nuclear pore, the exosome and the proteasome, or else are mitochondrial (Fig. 3, top). In contrast, many proteins with a faster than average turnover are involved in either mitosis, or other aspects of cell cycle regulation (Fig. 3, bottom). This includes protein components of the centromere, proteins with microtubule motor activity, proteins involved in cytoskeleton reorganization and proteins involved in chromatin assembly and condensation. We note that this study analyzed unsynchronized HeLa cells where only a minor fraction of the cells at any time point would be in mitosis.
Protein Turnover in Different Subcellular Compartments
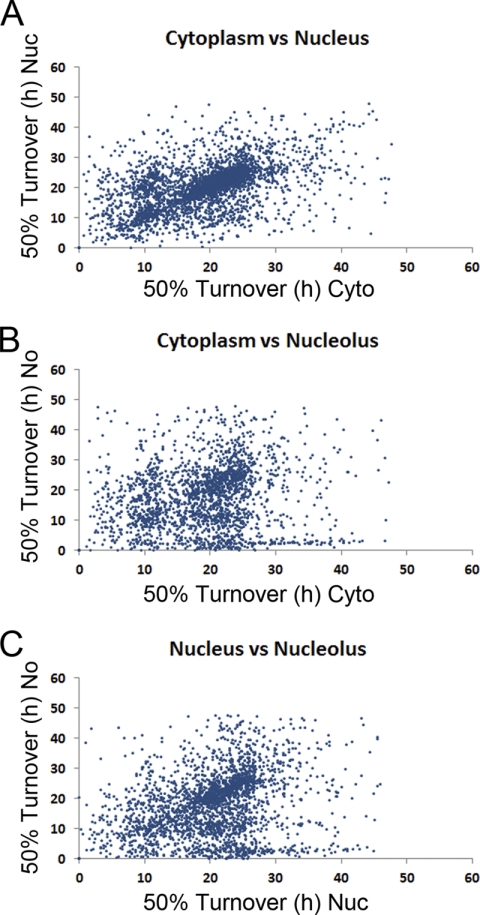
We have used the spatial proteomics approach (17, 18) combined with pulsed SILAC to measure the turnover of proteins in the separate cytoplasmic, nucleoplasmic, and nucleolar fractions. The turnover data for subcellular compartments are plotted against each other for comparison (Fig. 4). This shows that most proteins have a similar turnover rate in each compartment, particularly comparing nucleus and cytoplasm (Fig. 4A versus 4B and 4C). We performed correlation analyses between the different compartments and found that the Pearson correlation coefficient between the cytoplasm and the nucleus is 0.67, compared with 0.42 between the cytoplasm and the nucleolus and 0.50 between the nucleus and the nucleolus.
Fig. 4.
Protein turnover in subcellular compartments. The turnover data for subcellular compartments are plotted against each other to compare the 50% turnover values for each protein in the nucleus versus the cytoplasm (A), the nucleolus versus the cytoplasm (B) and the nucleolus versus the nucleus (C).
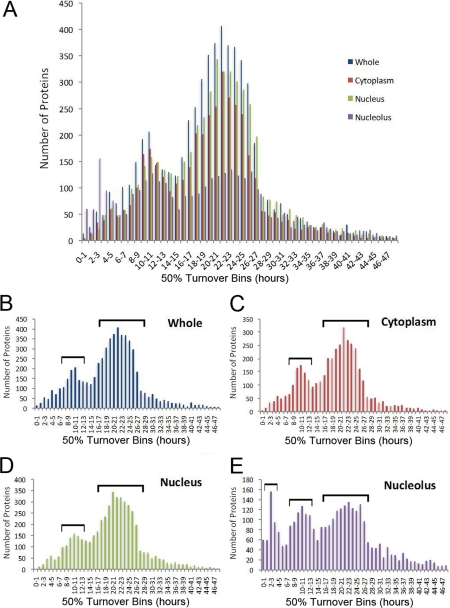
Protein turnover values of HeLa proteins follow an apparent bimodal distribution, with a major peak in the number of proteins with a 50% turnover value of ∼20 h, and a minor peak of ∼10 h (Figs. 5A, 5B and 5C). The similar distribution of protein turnover for the cytoplasm and the nucleoplasm (Fig. 5C and 5D), contrasts with the nucleolus, where there appears to be a third peak with a faster 50% turnover rate (<6 h), while the major peak is centered ∼22–23 h, slower than the whole cell mean turnover value of ∼20 h (Fig. 3E). The nucleolar proteins with the fastest turnover rate are predominantly ribosomal proteins. This may reflect the subset of proteins and more specialized functions occurring within the nucleolus as opposed to throughout the rest of the cell.
Fig. 5.
Distribution of protein turnover in subcellular compartments. A distribution plot with the number of proteins on the y axis and 50% turnover values (in bins of 1 h intervals) on the x axis for the whole cell (B), cytoplasmic (C), nuclear (D) or nucleolar (E) proteins, as well as an overlay of all four (A).
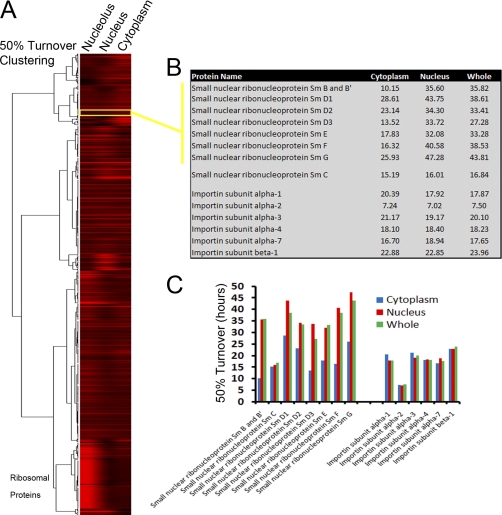
A clustering analysis grouped proteins with similar turnover values in either the cytoplasm, nucleoplasm or nucleoli, represented as a heat map with the protein clusters on the y axis and the subcellular compartments on the x axis (Fig. 6A). Most proteins showed similar turnover values in each compartment. Ribosomal proteins provide a clear example of a protein cluster with differing turnover values between compartments, i.e. fast turnover in the nucleolus (∼6 h), but slow turnover in the cytoplasm (>30 h), (Fig. 6A, bottom cluster). Other examples were identified where multiple subunits of the same multiprotein complex also show differential turnover values in one of the subcellular compartments. For example, Sm proteins, (components of the small nuclear ribonucleoprotein (snRNP) spliceosome subunits), showed faster turnover of ∼18 h in the cytoplasm, where snRNP proteins are assembled on snRNAs, compared with an average of ∼35 h in the nucleus, where the snRNPs function to splice pre-mRNAs (Figs. 6B and 6C). Interestingly, the Sm C protein, which is not part of the same complex as the other Sm subunits, did not show this difference in protein turnover between compartments. Other complexes with differences in subunit turnover values between compartments include the 26S proteasome, nuclear pore, T-complex, and RNA polymerase II. A common feature is that protein subunits have a faster turnover in the compartment where the complex assembles and are more stable in the compartment where the fully assembled complex functions.
Fig. 6.
Clustering analysis of protein turnover in subcellular compartments. A, A hierarchical clustering using the 50% turnover values for proteins in the cytoplasm, the nucleus, and the nucleolus is shown represented as a heat map. Fast turnover values are represented in red and slow turnover in black. B, A table showing the 50% turnover of the Sm proteins, i.e. subunits of the small nuclear ribonucleoprotein (snRNP) spliceosome and the Importin transport receptor proteins in the three subcellular compartments. C, Graphical representation of the 50% turnover value of each protein in the cytoplasm (blue), the nucleus (red) or the average for the whole cell (green), with the turnover on the y axis.
Protein Characteristics Related to Turnover
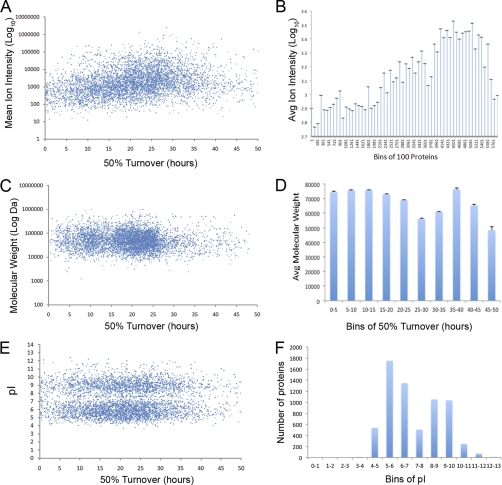
A range of protein properties and characteristics, including abundance, size, pI values, sequence motifs, and amino acid composition were analyzed for correlations with turnover (Figs. 6, 7, and supplementary Information). A positive correlation was detected between protein abundance, as measured from peptide intensity, and protein turnover values (Figs. 7A and 7B). This correlation was also recently observed in a study of protein turnover in mouse cells (28). While there is variation, higher abundance proteins generally had slower than average turnover rates (Fig. 7B). The corollary is that the time to turn over half of the total protein content of a HeLa cell is ∼15–20% longer than the mean turnover value of all proteins measured.
Fig. 7.
Protein characteristics related to turnover. A, Protein abundance was estimated from the averaged sum of ion intensities measured for every peptide in a protein and plotted on the y axis versus the turnover on the x axis. B, A distribution plot with the average log base 10 intensity on the y axis and bins of 100 proteins on the x axis, where proteins are sorted from the fastest turnover to the slowest turnover for the whole cell. C, The log base 10 of molecular weight (in Daltons) was plotted versus the protein turnover in the whole cell. D, A distribution plot of the average molecular weight in Daltons on the y axis and turnover (shown in 5 h bins) on the x axis. E, A comparison of the protein turnover on the x axis with isoelectric point on the y axis. F, A distribution plot of the number of proteins in each bin of isoelectric points.
In contrast with the positive correlation with abundance, there is minimal correlation between turnover and protein size (Figs. 7C and 7D). Comparison of predicted molecular weights deduced from amino acid sequences with measured 50% protein turnover values showed a Pearson correlation coefficient of –0.09 (Fig. 7B). It has been proposed that acidic proteins are degraded more rapidly than basic proteins (29). A comparison of the rate of protein turnover with isoelectric point, however, showed no correlation (Fig. 7E, Pearson correlation 0.009). We conclude that the bulk charge property of HeLa proteins is not a significant determinant of their stability. However, the analysis showed that nucleolar proteins have an inverse correlation between pI and protein turnover, with a Pearson correlation of –0.23. Thus, basic nucleolar proteins have a faster than average turnover (Fig. 5E, purple), likely because of the large number of basic ribosomal proteins in the nucleolus, which have a very fast turnover.
The presence of protein segments rich in proline, glutamic acid, serine, and threonine, (PEST sequences) are reported to affect degradation levels (30). We therefore analyzed the proteins whose turnover was measured for the presence and frequency of PEST motifs. While no simple correlation was observed between the presence of PEST sequences per se, and fast turnover values (supplemental Fig. S6), the average number of PEST regions found in proteins with faster than average turnover was ∼1, as compared with a PEST frequency of ∼0.5 for proteins with a slower than average turnover. We conclude that there is a positive relationship between the presence of PEST motifs and protein turnover in HeLa cells. However, the considerable variability in the 50% turnover values of specific proteins containing PEST motifs points to a complex relationship between sequence motifs and protein stability. It is likely that multiple structural and sequence elements, as well as abundance, localization, and the presence of interaction partners, can all affect the net stability of individual proteins.
Protein Turnover and the N-Terminal Amino Acid Rule
A previously characterized determinant of protein stability is the N-terminal amino acid of the mature protein, where the N-terminal amino acid is classified as either stabilizing, or destabilizing (31). Although for most proteins, methionine is the first amino acid encoded and translated, methionine aminopeptidase is thought to remove the N-terminal methionine when the second amino acid is either C, G, A, S, T, C, or P (32). Some mature proteins can also be generated by post-translational cleavage, resulting in different amino acids occurring at the N terminus. We used our empirical measurements of protein turnover rates to test whether the identity of the N- or C-terminal amino acid could affect the stability of full-length endogenous proteins in HeLa cells. The 50% turnover values were averaged for all proteins measured with each amino acid at either the first ten N-terminal positions (i.e. +1 to +10), or the last ten positions from the C terminus (supplemental Table S3). No significant differences were observed between mean protein turnover values according to the amino acid identity at either the first or last ten N-terminal, or C-terminal positions, respectively. We also made this comparison specifically for the 10% fastest and 10% slowest turnover proteins and also saw no correlation with amino acid identity at N- or C- termini (supplemental Table S3). We conclude that protein turnover for full-length, endogenous proteins in HeLa cells is not determined primarily by either N-terminal or C-terminal rules based upon amino acid identity.
The matrix of frequencies for each amino acid occurring at either the first ten N-terminal positions or last ten C-terminal positions in each protein identified was also determined and compared with the corresponding in silico prediction of amino acid frequencies for each ORF in the human genome. The resulting Pearson correlation coefficient of 0.99 shows that the sample of 6402 proteins for which 50% turnover values were measured have a near identical distribution of N- and C-terminal amino acid frequencies to the total translated human proteome (supplemental Table S4). We conclude that the subset of human proteins sampled in our study is thus highly representative of the total human proteome.
Viewer Description
We have implemented a database viewer within the PepTracker software environment to provide convenient access to these data through a web-based interface (http://www.lamondlab.com/turnover/). The application includes a search facility (supplemental Fig. S5) that allows users to search for a protein(s) of interest using protein name, description, gene as well as IPI or Uniprot identifiers. Proteins can be selected also using the interactive chart component on the home page, providing users with the ability to click either on chart items or the cell map to drill down into data from the cell fraction level to proteins and further to peptides. Right clicking a bar chart item accesses detailed data for the selected protein (supplemental Fig. S5C). The search result page documents also spatial proteomics localization data from human HCT116 cells, (17), as well as the localization data resulting from averaging the intensity values of the peptides identified in each subcellular compartment in this study (supplemental Fig. S5C). The viewer displays all peptides identified for a selected protein sequence, with different shading for each peptide reflecting differences in 50% turnover values. The curve fits are displayed showing degradation and synthesis rates for each protein, and showing the turnover rate of each protein in each subcellular compartment. Protein turnover for a selected protein is shown overlaid in red on scatter plots showing all proteins in the respective subcellular compartments in blue, providing a rapid overview of turnover rates between subcellular compartments.
DISCUSSION
We have developed a combined pulse-labeling, spatial proteomics and data analysis strategy to characterize the expression, localization, synthesis, degradation, and turnover rates of endogenously expressed, untagged human proteins in different subcellular compartments. Using SILAC and mass spectrometry, a total of 80,098 peptides from an estimated 8041 HeLa proteins were quantified, and their spatial distribution between the cytoplasm, nucleus, and nucleolus determined and visualized using PepTracker. To our knowledge, this study provides the first systematic, system wide quantitative analysis of proteome localization and turnover that has evaluated the properties of endogenous proteins in different subcellular compartments. The approach described, together with the software tools for data visualization and analysis, provides a basis for further systematic proteome-wide characterization of protein localization and turnover that can be compared between different cell types, cell cycle stages, physiological conditions, and genetic backgrounds.
Together with our collaborators, we previously used a heavy-light amino acid pulse SILAC protocol to measure protein turnover in HeLa cell nucleoli and compared the MS data with parallel studies on turnover of green fluorescent protein (GFP)-tagged nucleolar proteins using fluorescence microscopy (7). Similar heavy-light pulse SILAC approaches were used to study turnover of human A549 lung carcinoma cell proteins (33) and most recently to study also mouse NIH3T3 cells (28). In our present study we extend the pulse SILAC technique by analyzing a combination of heavy-medium pulsed cells and an equal amount of control, light cells at each time point. This offers advantages in terms of improved data analysis and statistical evaluation procedures. Overall, the pulse SILAC approach is useful for determining protein turnover because it allows measurement of endogenous proteins expressed at physiological levels while avoiding the need to treat cells with translation inhibitors. Techniques based on translation inhibition complicate the interpretation of protein turnover values as the effect of the inhibitors on cell physiology, which can in turn affect protein stability, must be taken into account (34).
Many large-scale, functional genomics studies characterize global gene expression levels, either in different cell types and/or under a range of growth conditions, by measuring differences in mRNA expression levels. This either involves microarray technology, or, more recently, high-throughput RNA sequencing. However, quantitative mRNA expression data alone are not sufficient to reliably document gene expression at the proteome level. Similar mRNA expression levels can be accompanied by a wide range (up to 20-fold difference) in the corresponding abundance levels of the proteins encoded by these mRNAs (3). We observe only weak correlations (Pearson correlation coefficients ∼0.2) comparing our estimates of HeLa protein expression levels with publicly available HeLa mRNA expression data (ArrayExpress (EBI), data not shown). This overall poor correlation between cognate mRNA and protein expression levels likely reflects differences both in rates of mRNA translation and in protein turnover, but may also, at least in part, result from noisy microarray measurements and variability of HeLa cell batches. It is important to note that many proteins can differ substantially in their in vivo half-lives, regardless of how fast they are synthesized (35). This underlines the importance of making direct measurements of endogenous cell proteins, including the high-throughput analysis of protein turnover, to fully evaluate gene expression responses and accurately determine factors and mechanisms regulating intracellular protein abundance.
In silico translation of the human genome shows the proteins identified in this study are highly representative of the human proteome. Several lines of evidence also argue that the MS-based proteomics approach used here is robust and reproducible. First, a strong positive correlation (Pearson correlation coefficient ∼0.73) was observed between the present data and the smaller data set from our previous pulse SILAC analysis of HeLa nucleolar protein turnover (7), despite the lower number of peptides identified for each protein in that case. This demonstrates that, at least when comparing the same cell line, biological replicates produce similar results. Second, we have confirmed that values from technical replicates are reproducible by evaluating data obtained from separate MS analysis and quantitation of the same protein samples using different mass spectrometers. Third, we note that there was a strong positive correlation between the localization and turnover values obtained for most shared subunits of common multiprotein complexes, consistent with proteins in the same complex having similar biological properties. Most peptides in the same protein also produced similar values.
Our approach differs in several aspects from most previous studies on global protein turnover (28, 33, 34, 36, 37). First, we simultaneously determine not only net protein turnover, but also both protein degradation and synthesis rates. This provides additional information on the protein properties and allows calculation of protein turnover using two separate methods and statistical evaluation of the accuracy of the turnover values for each protein. Second, we characterize protein turnover not only for the global protein population in whole cells, but also for proteins in separate subcellular fractions. This spatial information recognizes that separate pools of protein with distinct properties can exist in different subcellular locations. Inevitably, analysis of whole cell extracts measure average values for the protein population and will not identify cases where the same protein can be present in more than one complex with different turnover rates, as revealed in this study. Third, our measurements are made on endogenous, untagged cell proteins and not based upon analysis of over-expressed, tagged fusion constructs. Either transient, or stable, overexpression of tagged fusion proteins may affect their turnover properties, both through changing the protein structure and by altering their abundance and stoichiometry relative to interaction partners.
A comparison of the protein turnover values reported here with the corresponding turnover or half-life values reported for the same proteins in previous large-scale studies showed major differences. Thus, there appeared to be a near random correlation with the values reported in two of these studies (33, 37), and only a partial positive correlation (Pearson correlation coefficient ∼0.2) with the data of Yen et al. (36). We note that cross comparison of the data sets from each of these previous studies also shows mostly random correlations between them (data not shown). However, we found a stronger correlation (Pearson correlation coefficient of 0.34) between our data and a recent study also using a pulse-SILAC method to analyze protein turnover in mouse NIH 3T3 cells (28).
The lack of correlation between many previous high-throughput studies on human cells and our present data is not simply explained by variation in the quality of our data. Even focusing on the subset of proteins in our study with the highest quality measurements did not significantly improve the degree of correlation. Thus, considering only the ∼25% of proteins for which we quantitated at least 20 separate peptides with optimal chi2 curve fitting did not increase the positive correlation with the other data sets. We conclude that we have generated a data set for endogenous human proteins that is distinct from previous studies and, considering the overall stringent data evaluation employed, argue that the lack of agreement in protein turnover values between our data and previous large-scale studies is not primarily reflecting data quality issues in our measurements.
This surprising situation where apparently each separate high-throughput analysis of protein turnover produces different results could have multiple explanations. Two of the previous studies specifically analyzed the half-lives of over-expressed, GFP-tagged fusion proteins (36, 37). We anticipate that the resulting fusion protein turnover values may differ from the turnover values reported here for the corresponding endogenous proteins expressed at physiological levels. It is also important to note, however, that there is no expectation that different cell lines growing under different culture conditions should show identical protein turnover values, as observed from the differences in turnover measured between A549 cells (33), NIH 3T3 cells (28), and HeLa cells (this study). It will be interesting, therefore, to carefully evaluate differences in protein turnover values between cell lines and growth conditions using the same stringent methodologies for all measurements. Indeed, although most homologous proteins showed a similar trend when comparing turnover values from our study to those found in NIH 3T3 cells (28), it is interesting to note that a few proteins showed a dramatic difference in protein turnover between the two cell lines, indicating that specific protein degradation might be either cell type specific, or species-specific, or both.
An interesting general feature from this study is that protein subunits from multiprotein complexes show faster turnover as free proteins, prior to complex assembly. This is exemplified by the ribosomal proteins, which have a 50% turnover of ∼6 h in the nucleus, where the protein pool includes free, unassembled ribosomal proteins (7). In contrast, ribosomal proteins are very stable in the cytoplasm, with turnover of over 30 h, where they accumulate only after assembly into a ribosome subunit. An important corollary is that a large increase in the expression of a specific protein, as often occurs upon either transient or stable over-expression of tagged proteins, may change drastically its turnover in comparison with the endogenous counterpart. The measured turnover values of interaction partners of the over-expressed factor and of other proteins may also be altered. We propose that this could account for much of the difference between the turnover values we report here for endogenous proteins and the faster turnover measured using fluorescent protein-tagged proteins. For example, Yen et al., reported that the turnover rates of over 8000 GFP-tagged human proteins showed a bimodal pattern, where the average turnover values were measured as 30 min and 2 h, respectively (36). Here we also found that protein turnover followed a bimodal distribution, but with slower turnover values of ∼20 h, close to the HeLa cell division rate and a minor peak with a turnover value of ∼10 h.
The data indicate that the bulk of HeLa cell proteins may be turned over passively during normal cell growth and are consistent with the mean turnover rate reflecting approximate doubling of the amount of proteins as the cell divides and hence doubles its protein content. However, a subset of proteins show faster turnover, suggesting they may be directly targeted for degradation. The similar distribution of protein turnover values seen for the cytoplasm and the nucleoplasm likely reflects the high level of protein shuttling between these compartments. This contrasts with the nucleolus, where a distinct group of proteins show fast turnover (<6 h), mostly corresponding to ribosomal proteins. Interestingly, recent studies point to a role for the accumulation of specific free ribosomal proteins in the nucleus in signaling mechanisms involved in stress responses and growth control (38), suggesting that the control of ribosomal protein stability in the nucleus is involved in biological regulation.
Proteins with the slowest turnover have a wide range of functions, but are commonly present either in large, abundant and stable protein complexes, such as ribosome and spliceosome subunits, RNA polymerases, the nuclear pore, the exosome and the proteasome, or else are found inside mitochondria. Interestingly, with almost all of these slow turnover proteins, we note that the turnover rate of each subunit was significantly slower in one subcellular compartment, correlating with the location where they exert their function. These observations suggest a general assembly strategy whereby cells produce an excess of subunits in order to favor complex formation, but carry out this assembly in a compartment separate to the eventual main site of function. This avoids the need to tightly coregulate transcription, processing, transport and translation of the mRNAs encoding different protein subunits in eukaryotes where genes are not organized in operons and not co-transcribed and translated. Any excess protein subunits produced will simply be degraded in the assembly compartment. This model explains the differential stability of ribosomal proteins between the nucleus, where they are assembled with RNA, and the cytoplasm where they function to translate mRNA and conversely, the higher stability of snRNP proteins in the nucleus, where they function in pre-mRNA splicing, as opposed to in the cytoplasm, where they assemble on snRNAs.
We envisage in future that this general approach for characterizing protein turnover and associated protein properties can be extended in several ways. It is possible to expand the subcellular fractionation strategy for example and thereby obtain higher resolution spatial information regarding the subcellular distribution of the proteome and how this correlates with protein structure, isoforms and PTM patterns. Our present study has not distinguished effects on the proteome of cells growing at different stages of the cell cycle. However, specific examples are already known where either protein stability, or subcellular localization, can alter as cells progress through interphase and mitosis. Work is in progress therefore to carry out systematic, proteome-wide analyses of how protein properties, including turnover rates and subcellular localization patterns, vary as a function of cell cycle progression, providing a detailed quantitative annotation of the human proteome in both time and space. None of the protein properties discussed above represent “absolute” values, and it is to be expected that rates of protein turnover, localization patterns, interaction partners and PTMs will vary considerably between different cell lines, under different growth conditions and in response to drugs or other external stimuli. Specific mutations, which may be associated with either oncogenic transformation or genetic disease, can also alter these protein properties. The development and integration of many large-scale, quantitative proteomic data sets of the sort described here thus offers a promising future direction for expanding the functional annotation of the human genome, and the genomes of other model organisms, and for the discovery of new biological regulatory mechanisms.
Acknowledgments
AIL is a Wellcome Trust Principal Research Fellow. Yasmeen Ahmad is supported by a BBSRC PhD studentship.
Footnotes
* This work was supported in part by the European Commission's FP7 (GA HEALTH-F4-2008-201648/PROSPECTS) (www.prospects-fp7.eu/), by RASOR (Radical Solutions for Researching the Proteome), and by a Wellcome Trust program grant to AIL (073980/Z/03/Z).
1 The abbreviations used are:
SILAC
stable isotope labeling with amino acids in cell culture
LC-MS/MS
liquid chromatography-tandem MS
HPLC
high performance liquid chromatography
IPI
International Protein Index
snRNP
small nuclear ribonucleoprotein.
REFERENCES
- 1.Ohsumi Y. (2006) Protein turnover. IUBMB Life 58, 363–369 [DOI] [PubMed] [Google Scholar]
- 2.Lundberg E., Fagerberg L., Klevebring D., Matic I., Geiger T., Cox J., Algenäs C., Lundeberg J., Mann M., Uhlen M. (2010) Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 6, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gygi S. P., Rochon Y., Franza B. R., Aebersold R. (1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walther T. C., Mann M. (2010) Mass spectrometry-based proteomics in cell biology. J. Cell Biol. 190, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clague M. J., Urbé S. (2010) Ubiquitin: same molecule, different degradation pathways. Cell 143, 682–685 [DOI] [PubMed] [Google Scholar]
- 6.Lane D., Levine A. (2010) p53 Research: the past thirty years and the next thirty years. Cold Spring Harb. Perspect. Biol. 2, a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garlick P. J., Millward D. J. (1972) An appraisal of techniques for the determination of protein turnover in vivo. Proc. Nutr Soc. 31, 249–255 [DOI] [PubMed] [Google Scholar]
- 9.Doherty M. K., Whitehead C., McCormack H., Gaskell S. J., Beynon R. J. (2005) Proteome dynamics in complex organisms: using stable isotopes to monitor individual protein turnover rates. Proteomics 5, 522–533 [DOI] [PubMed] [Google Scholar]
- 10.Schwanhäusser B., Gossen M., Dittmar G., Selbach M. (2009) Global analysis of cellular protein translation by pulsed SILAC. Proteomics 9, 205–209 [DOI] [PubMed] [Google Scholar]
- 11.Pratt J. M., Petty J., Riba-Garcia I., Robertson D. H., Gaskell S. J., Oliver S. G., Beynon R. J. (2002) Dynamics of protein turnover, a missing dimension in proteomics. Mol. Cell. Proteomics 1, 579–591 [DOI] [PubMed] [Google Scholar]
- 12.Milner E., Barnea E., Beer I., Admon A. (2006) The turnover kinetics of major histocompatibility complex peptides of human cancer cells. Mol. Cell. Proteomics 5, 357–365 [DOI] [PubMed] [Google Scholar]
- 13.Ong S. E., Kratchmarova I., Mann M. (2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2, 173–181 [DOI] [PubMed] [Google Scholar]
- 14.Mann M. (2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 15.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 16.Ohta S., Bukowski-Wills J. C., Sanchez-Pulido L., Alves Fde L., Wood L., Chen Z. A., Platani M., Fischer L., Hudson D. F., Ponting C. P., Fukagawa T., Earnshaw W. C., Rappsilber J. (2010) The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142, 810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boisvert F. M., Lam Y. W., Lamont D., Lamond A. I. (2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell. Proteomics 9, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boisvert F. M., Lamond A. I. (2010) p53-Dependent subcellular proteome localization following DNA damage. Proteomics 10, 4087–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y. W., Steen H., Mann M., Lamond A. I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11 [DOI] [PubMed] [Google Scholar]
- 20.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 21.Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 22.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 23.Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 24.Carrillo B., Yanofsky C., Laboissiere S., Nadon R., Kearney R. E. (2010) Methods for combining peptide intensities to estimate relative protein abundance. Bioinformatics 26, 98–103 [DOI] [PubMed] [Google Scholar]
- 25.Davies D. D., Humphrey T. J. (1978) Amino Acid recycling in relation to protein turnover. Plant Physiol. 61, 54–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 27.Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 28.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 29.Dice J. F., Goldberg A. L. (1975) Relationship between in vivo degradative rates and isoelectric points of proteins. Proc. Natl. Acad. Sci. U.S.A. 72, 3893–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers S., Wells R., Rechsteiner M. (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234, 364–368 [DOI] [PubMed] [Google Scholar]
- 31.Varshavsky A. (1992) The N-end rule. Cell 69, 725–735 [DOI] [PubMed] [Google Scholar]
- 32.Hu R. G., Brower C. S., Wang H., Davydov I. V., Sheng J., Zhou J., Kwon Y. T., Varshavsky A. (2006) Arginyltransferase, its specificity, putative substrates, bidirectional promoter, and splicing-derived isoforms. J. Biol. Chem. 281, 32559–32573 [DOI] [PubMed] [Google Scholar]
- 33.Doherty M. K., Hammond D. E., Clague M. J., Gaskell S. J., Beynon R. J. (2009) Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome Res. 8, 104–112 [DOI] [PubMed] [Google Scholar]
- 34.Belle A., Tanay A., Bitincka L., Shamir R., O'Shea E. K. (2006) Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. U.S.A. 103, 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenbaum D., Colangelo C., Williams K., Gerstein M. (2003) Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yen H. C., Xu Q., Chou D. M., Zhao Z., Elledge S. J. (2008) Global protein stability profiling in mammalian cells. Science 322, 918–923 [DOI] [PubMed] [Google Scholar]
- 37.Eden E., Geva-Zatorsky N., Issaeva I., Cohen A., Dekel E., Danon T., Cohen L., Mayo A., Alon U. (2011) Proteome half-life dynamics in living human cells. Science 331, 764–768 [DOI] [PubMed] [Google Scholar]
- 38.Sundqvist A., Liu G., Mirsaliotis A., Xirodimas D. P. (2009) Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 10, 1132–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]