Development of Additional Selectable Markers for the Halophilic Archaeon Haloferax volcanii Based on the leuB and trpA Genes (original) (raw)
Abstract
Since most archaea are extremophilic and difficult to cultivate, our current knowledge of their biology is confined largely to comparative genomics and biochemistry. Haloferax volcanii offers great promise as a model organism for archaeal genetics, but until now there has been a lack of a wide variety of selectable markers for this organism. We describe here isolation of H. volcanii leuB and trpA genes encoding 3-isopropylmalate dehydrogenase and tryptophan synthase, respectively, and development of these genes as a positive selection system. Δ_leuB_ and Δ_trpA_ mutants were constructed in a variety of genetic backgrounds and were shown to be auxotrophic for leucine and tryptophan, respectively. We constructed both integrative and replicative plasmids carrying the leuB or trpA gene under control of a constitutive promoter. The use of these selectable markers in deletion of the lhr gene of H. volcanii is described.
Less than 25 years ago, the archaea were virtually unknown. We now recognize that these organisms represent one of the fundamental domains of life (32) and constitute a significant fraction of the total biomass (9). Acceptance of the distinct status of the archaea has been largely due to genome sequencing projects (6). The data from the sequenced genomes have revealed that in spite of their prokaryotic morphology, the archaea have numerous similarities with eukaryotes (5), particularly in enzymes involved in core processes such as transcription (2), translation (10), and DNA replication (23). As the archaeal transcription and replication systems are considerably less complex than those found in eukaryotes, they are more amenable to analysis. Notwithstanding this sequence similarity, the archaea have a unique identity (12), which is best exemplified by novel enzymes such as the Holliday junction resolvase Hjc (19).
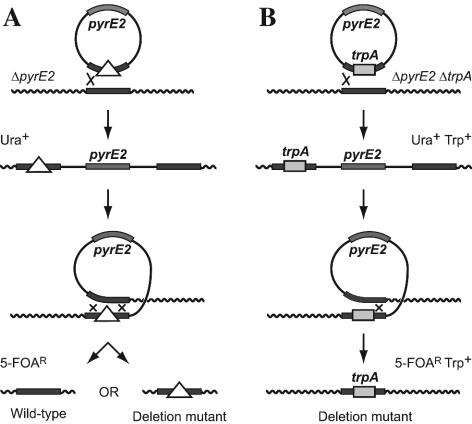
In order to realize the true potential of the archaea, both as stripped-down models to dissect more complex eukaryotic systems and as a source of novel enzymes, it is essential to harness the power of genetics to underpin advances in biochemistry and genomics. Haloferax volcanii is an obligate halophile (25) that is genetically stable and grows aerobically in both complex and minimal media (24). It has some of the best genetic tools among the archaea, including a transformation system (8), reporter genes (17), shuttle vectors with antibiotic resistance (14, 16, 22), auxotrophic markers (26), and a recently developed gene knockout system based on the pyrE2 gene (3) (Fig. 1A), which encodes orotate phosphoribosyl transferase and is involved in uracil biosynthesis.
FIG. 1.
Gene knockout system based on the pyrE2 gene. (A) A plasmid carrying the pyrE2 marker and flanking sequences of the gene to be deleted is used to transform a Δ_pyrE2 H. volcanii_ strain to uracil prototrophy. Here, the crossover used to integrate the plasmid (pop-in) has occurred to the left of the deletion. Subsequent loss of the plasmid by intrachromosomal crossing over can occur on the left of the deletion, restoring the gene to wild type, or on the right of the deletion, resulting in the desired mutant. In either case the cell is rendered auxotrophic for uracil and is therefore resistant to 5-FOA by virtue of its inability to convert this compound to the toxic analog 5-fluorouracil. (B) The gene is replaced with the trpA marker, and the plasmid is used to transform a Δ_pyrE2_ Δ_trpA H. volcanii_ strain to prototrophy for uracil and tryptophan. Loss of the plasmid by crossing over on the right of the deletion, resulting in a _trpA_-marked mutant, can be selected in one step.
Since the number of selectable genetic markers in H. volcanii is still limited, we wished to supplement it with additional markers. We describe here the isolation of H. volcanii leuB and trpA genes encoding 3-isopropylmalate dehydrogenase and tryptophan synthase, respectively. Deletion of leuB, a homologue of the widely used Saccharomyces cerevisiae LEU2 gene (1), confers leucine auxotrophy in minimal medium. Deletion of the trpA gene (20) confers tryptophan auxotrophy in minimal or Casamino Acids medium. The trpA marker is of particular value when it is used with the pyrE2 gene knockout system, as it allows direct selection for deleterious mutations that are otherwise difficult to recover (Fig. 1B). Here we describe the use of this system for deletion of the H. volcanii homologue of the Escherichia coli lhr gene (28). The leuB and trpA markers were combined with a deletion of the hdrB gene (26), conferring thymidine auxotrophy in rich (yeast extract) medium, and both integrating and shuttle plasmid vectors in which pyrE2, leuB, trpA, and hdrB were used as selectable markers were generated.
MATERIALS AND METHODS
Unless stated otherwise, chemicals were obtained from Sigma and restriction endonucleases were obtained from New England Biolabs.
Strains and culture conditions.
The H. volcanii strains used are shown in Table 1 and were routinely grown in rich medium (Hv-YPC) containing (per liter) 144 g of NaCl, 21 g of MgSO4 · 7H2O, 18 g of MgCl2 · 6H2O, 4.2 g of KCl, and 12 mM Tris HCl (pH 7.5). For solid media, agar (Difco) was added at a concentration of 15 g per liter and was dissolved by heating the medium in a microwave oven. Yeast extract (0.5%, wt/vol; Difco), 0.1% (wt/vol) peptone (Oxoid), and 0.1% (wt/vol) Casamino Acids (Difco) were added, and the medium was autoclaved. After cooling, CaCl2 was added to a final concentration of 3 mM. When required, novobiocin was added to a concentration of 2 μg/ml, mevinolin was added to a concentration of 4 μg/ml, and thymidine was added to a concentration of 40 μg/ml. Casamino Acids medium (Hv-Ca) was made in a similar manner, except that yeast extract and peptone were omitted and Casamino Acids was added to a final concentration of 0.5% (wt/vol). When required, thymidine or hypoxanthine was added at a concentration of 40 μg/ml, and tryptophan or uracil was added at a concentration of 50 μg/ml; for pop-out selection medium, 5-fluoroorotic acid (5-FOA) was added to a concentration of 50 μg/ml and uracil was added to a concentration of 10 μg/ml. Minimal medium (Hv-Min) contained the same concentration of salts as Hv-YPC, except that Tris HCl (pH 7.5) was added to a concentration of 42 mM. After autoclaving and cooling, 4.25 ml of a sodium dl-lactate solution (60%, wt/vol), 3.83 g of disodium succinic acid · 6H2O, 0.25 ml of glycerol, 5 ml of a 1 M NH4Cl solution, 6 ml of a 0.5 M CaCl2 solution, 2 ml of 0.5 M potassium phosphate buffer (pH 7.5), 1 ml of a trace element solution (24), 0.8 mg of thiamine, and 0.1 mg of biotin were added per liter of Hv-Min. When required, thymidine or hypoxanthine was added to a concentration of 40 μg/ml, and leucine, tryptophan, uracil, methionine, glycine, or pantothenic acid was added to a concentration of 50 μg/ml.
TABLE 1.
H. volcanii strains used
| Strain | Background | Derivation or referencea | Genotype and/or phenotype |
|---|---|---|---|
| DS70 | (DS70) | 31 | |
| WR340 | WFD11 | 3 | His− |
| WR480 | WFD11 | 3 | His− Δ_pyrE2_ |
| H18 | DS70 | DS70, pGB68 pop-in | pyrE2+::[Δ_pyrE2_ NovR] |
| H23 | DS70 | DS70, pTA70 pop-in | leuB+::[Δ_leuB_ MevR] |
| H26 | DS70 | H18 pop-out | Δ_pyrE2_ |
| H30 | WFD11 | WR480, pTA73 pop-in | His− Δ_pyrE2 leuB_+::[Δ_leuB pyrE2_+] |
| H37 | DS70 | H23 pop-out | Δ_leuB_ |
| H40 | WFD11 | H30 pop-out | His− Δ_pyrE2_ Δ_leuB_ |
| H42 | WFD11 | WR340, pTA93 pop-in | His−trpA+::[Δ_trpA_ MevR] |
| H43 | DS70 | DS70, pTA93 pop-in | trpA+::[Δ_trpA_ MevR] |
| H45 | WFD11 | WR480, pTA95 pop-in | His− Δ_pyrE2 trpA_+::[Δ_trpA pyrE2_+] |
| H47 | DS70 | H26, pTA95 pop-in | Δ_pyrE2 trpA_+::[Δ_trpA pyrE2_+] |
| H52 | WFD11 | H45 pop-out | His− Δ_pyrE2_ Δ_trpA_ |
| H53 | DS70 | H47 pop-out | Δ_pyrE2_ Δ_trpA_ |
| H60 | DS70 | H26, pTA73 pop-in | Δ_pyrE2 leuB_+::[Δ_leuB pyrE2_+] |
| H66 | DS70 | H60 pop-out | Δ_pyrE2_ Δ_leuB_ |
| H76 | WFD11 | H42 pop-out | His− Δ_trpA_ |
| H77 | DS70 | H43 pop-out | Δ_trpA_ |
| H90 | DS70 | H26, pTA155 pop-in | Δ_pyrE2 hdrB_+::[Δ_hdrB pyrE2_+] |
| H91 | DS70 | H53, pTA155 pop-in | Δ_pyrE2_ Δ_trpA hdrB_+::[Δ_hdrB pyrE2_+] |
| H92 | DS70 | H66, pTA155 pop-in | Δ_pyrE2_ Δ_leuB hdrB_+::[Δ_hdrB pyrE2_+] |
| H98 | DS70 | H90 pop-out | Δ_pyrE2_ Δ_hdrB_ |
| H99 | DS70 | H91 pop-out | Δ_pyrE2_ Δ_trpA_ Δ_hdrB_ |
| H100 | DS70 | H92 pop-out | Δ_pyrE2_ Δ_leuB_ Δ_hdrB_ |
| H107 | DS70 | H26, pTA166 pop-in | Δ_pyrE2 lhr_+::[Δ_lhr pyrE2_+] |
| H108 | DS70 | H53, pTA166 pop-in | Δ_pyrE2_ Δ_trpA lhr_+::[Δ_lhr pyrE2_+] |
| H109 | DS70 | H53, pTA172 | Δ_pyrE2_ Δ_trpA lhr_+::[Δ_lhr_::trpA+pyrE2+] |
| H111 | DS70 | H53, pTA73 pop-in | Δ_pyrE2_ Δ_trpA leuB_+::[Δ_leuB pyrE2_+] |
| H119 | DS70 | H111 pop-out | Δ_pyrE2_ Δ_trpA_ Δ_leuB_ |
| H120 | DS70 | H107 pop-out | Δ_pyrE2_ Δ_lhr_ |
| H121 | DS70 | H108 pop-out | Δ_pyrE2_ Δ_trpA_ Δ_lhr_ |
| H122 | DS70 | H109 pop-out | Δ_pyrE2_ Δ_trpA_ Δ_lhr_::trpA+ |
| H126 | DS70 | H119, pTA155 pop-in | Δ_pyrE2_ Δ_trpA_ Δ_leuB hdrB_+::[Δ_hdrB pyrE2_+] |
| H133 | DS70 | H126 pop-out | Δ_pyrE2_ Δ_trpA_ Δ_leuB_ Δ_hdrB_ |
E. coli strains XL1-Blue MRF′ (Δ_mcrA183_ Δ_mcrCB-hsdSMR-mrr173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac_ [F′ proAB lacIq _Z_ΔM15 Tn_10_]) and GM121 (F− dam-3 dcm-6 ara-14 fhuA31 galK2 galT22 hdsR3 lacY1 leu-6 thi-1 thr-1 tsx-78) were grown in Luria-Bertani medium containing 100 μg of ampicillin per ml. The latter strain was used to prepare unmethylated plasmid DNA for efficient transformation of H. volcanii (18).
Transformation procedures.
Transformation of H. volcanii was carried out by using polyethylene glycol 600 as described previously (8). The media used for transformation were prepared as described above, except that sucrose was added to a concentration of 15% (wt/vol). E. coli was transformed by using a standard electroporation protocol (29).
Molecular genetic methods.
Restriction endonuclease digestion, agarose gel electrophoresis, Southern blot analysis, and molecular cloning techniques were performed by using standard procedures (29). Probes were generated by PCR by using the provisional H. volcanii genome sequence (University of Scranton, Scanton, Pa.) and the primers listed in Table 2.
TABLE 2.
Oligonucleotide primers used
| Primer | Sequence (5′-3′)a | Relevant propertiesb |
|---|---|---|
| HvLeuF | CGCCGGCGACCACGTCAAAGAAGA | leuB probe, forward primer |
| HvLeuR | AGCAGCATCGCCGCGGACAGAATC | leuB probe, reverse primer |
| TrpAF2 | CGCCGAGGGGCCGACCATCC | trpA probe, forward primer |
| TrpAR | CGTTGCGACGCGCCCGCTACC | trpA probe, reverse primer |
| dLeu5F | GCGTTCAGCACGAATTCCGCCGCCGGGATGACCT | leuB deletion, upstream internal primer, _Eco_RI deletion site |
| dLeu5R | CGCGGGATCCGTCAACCCCGACGAGACCACCTACGA | leuB deletion, upstream external primer, _Bam_HI cloning site |
| dLeu3F | GCACGGATCCGCGGGCCGTTGTGATTGAGT | leuB deletion, downstream external primer, _Bam_HI cloning site |
| dLeu3R | GGCGGAATTCGTTTCGAACGCGCCCGTTTTCGTTTCTGAT | leuB deletion, downstream internal primer, _Eco_RI deletion site |
| dTrp5F | GCTCTAGAACGCGCTCGGGCAGGTCTTACTGG | trpA deletion, upstream external primer, _Xba_I cloning site |
| dTrp5R | GGACGAATTCCGGGCCGTCGGAGAAGG | trpA deletion, upstream internal primer, _Eco_RI deletion site |
| dTrp3F2 | CGAACTCGAATTCGGTGCGGTAGCG | trpA deletion, downstream internal primer, _Eco_RI deletion site |
| dTrp3R | CCGGTGAGTCTCTAGACGTTTTCGTCCG | trpA deletion, downstream external primer, _Xba_I cloning site |
| TrpPci | GCCTGACATGTCGCTCGAAGACGCC | trpA coding sequence, forward primer, _Pci_I site |
| TrpXba | GGGTTCTAGAGCAGTTATGTGCGTTCC | trpA coding sequence, reverse primer, _Xba_I site |
| LeuBsp | GCCCTACGTTCATGACTGAGGAAATCG | leuB coding sequence, forward primer, _Bsp_HI site |
| LeuXba | CGGGTCGCTCTAGATCAGAGTCGGTCG | leuB coding sequence, reverse primer, _Xba_I site |
| LhrF2 | GAAGCTGAAGGCGGGCGAGTTACG | lhr probe, forward primer |
| LhrR2 | ATGGCGGCGAGGTTCAGTTTGTCT | lhr probe, reverse primer |
| dHdrBF2 | CCCGATCTAGAGCCGGCTGGTCATC | hdrB deletion, upstream external primer, _Xba_I cloning site |
| dHdrB5R | CCCAGAAAGCTGCTAGCCGCTCATTCG | hdrB deletion, upstream internal primer, _Nhe_I deletion site |
| dHdrB3F | CTCGGGCTAGCGGGAGTACAAAATCGTC | hdrB deletion, downstream internal primer, _Nhe_I deletion site |
| dHdrB3R | GCCAAGCTCGAAATTAACCCTCAC | hdrB deletion, downstream external primer (_Xba_I site used) |
| dLhr5F | GAGCGCGCGTAATACGACTCACT | lhr deletion, upstream external primer (_Xho_I site used) |
| dLhr5R | GCGCGTCGCGGCCGCAATCAACGACG | lhr deletion, upstream internal primer, _Not_I deletion site |
| dLhr3F | GCGAGCGGCCGCGCCGGGTCATTACC | lhr deletion, downstream internal primer, _Not_I deletion site |
| dLhr3R | CGCGCAATTAACCCTCACTAAAGGG | lhr deletion, downstream external primer (_Xho_I site used) |
| HdrBsp | TGGCCTCATGAGCGGCGAGGAGC | hdrB coding sequence, forward primer, _Bsp_HI site |
| HdrXba | CTCCCACTCTCTAGAGTTACTCATCGG | hdrB coding sequence, reverse primer, _Xba_I site |
Isolation of H. volcanii total genomic DNA.
One milliliter of a saturated culture (grown in Hv-YPC broth) was centrifuged at 3,300 × g for 8 min and resuspended in 200 μl of 1 M NaCl-20 mM Tris HCl (pH 7.5). Then 200 μl of 100 mM EDTA (pH 8.0)-0.2% (wt/vol) SDS was added to lyse the cells, followed by 1 ml of ethanol. The DNA was spooled out of solution onto a capillary, washed twice with ethanol, and resuspended in 500 μl of TE (10 mM Tris HCl, 1 mM EDTA; pH 7.5). The DNA was precipitated with isopropanol, washed thoroughly with 70% ethanol, and resuspended in 100 μl of TE containing 0.1 mg of RNase A per ml.
Plasmid construction.
The plasmids used are shown in Table 3. Apart from the plasmids shown in Fig. 5 (see below), plasmids were constructed as described in the Results. The PCR primers used for generation of deletion constructs are listed in Table 2. Sequence files are available on request.
TABLE 3.
Plasmids used
| Plasmid | Relevant properties | Source or reference(s) |
|---|---|---|
| pBluescript II SK+ | Standard cloning vector | Stratagene |
| pD4 | pBluescript KS with H. volcanii 3,566-bp _Mbo_I-_Hin_dIII fragment containing hdrB gene | 26 |
| pGB68 | pBR322 with NovR and flanking sequences of pyrE2 | 3 |
| pGB70 | pUC19 with pyrE2 under ferredoxin promoter | 3 |
| pMDS99 | Shuttle vector based on pOK12 with pHV2 replication origin and MevR from Haloarcula hispanica | 31 |
| pWL102 | Shuttle vector based on pAT153 with pHV2 replication origin and MevR from H. volcanii | 7, 22 |
| pTA44 | pBluescript II with H. volcanii 4,162-bp _Bss_HII fragment containing leuB gene | This study |
| pTA49 | pBluescript II with H. volcanii 3,676-bp _Sau_3AI fragment containing trpA gene | This study |
| pTA65 | pBluescript II with _Bam_HI PCR fragment containing flanking regions of leuB | This study |
| pTA70 | pTA65 with _Not_I fragment from pMDS99 containing MevR | This study |
| pTA73 | pGB70 with _Xba_I-Hin_dIII Δ_leuB fragment from pTA65 | This study |
| pTA92 | pBluescript II with _Xba_I PCR fragment containing flanking regions of trpA | This study |
| pTA93 | pTA92 with _Not_I fragment from pMDS99 containing MevR | This study |
| pTA95 | pGB70 with Xba_I Δ_trpA fragment from pTA92 | This study |
| pTA131 | pBluescript II with _Bam_HI-_Xba_I fragment from pGB70 containing pyrE2 under ferredoxin promoter | This study |
| pTA132 | pBluescript II with PCR fragment containing trpA under ferredoxin promoter | This study |
| pTA133 | pBluescript II with PCR fragment containing leuB under ferredoxin promoter | This study |
| pTA150 | pBluescript II with H. volcanii 3,974-bp _Xho_I-_Nru_I fragment containing lhr gene | This study |
| pTA155 | pTA131 with _Hin_dIII-_Xba_I PCR fragment containing flanking regions of hdrB | This study |
| pTA166 | pTA131 with _Xho_I-_Spe_I PCR fragment containing flanking regions of lhr | This study |
| pTA172 | pTA166 with PCR fragment containing trpA under ferredoxin promoter, inserted at site of lhr deletion | This study |
| pTA192 | pBluescript II with PCR fragment containing hdrB under ferredoxin promoter | This study |
| pTA230 | pTA131 with _Nco_I-_Hin_dIII fragment from pWL102 containing pHV2 replication origin | This study |
| pTA231 | pTA132 with _Nco_I-_Hin_dIII fragment from pWL102 containing pHV2 replication origin | This study |
| pTA232 | pTA133 with _Nco_I-_Hin_dIII fragment from pWL102 containing pHV2 replication origin | This study |
| pTA233 | pTA192 with _Nco_I-_Hin_dIII fragment from pWL102 containing pHV2 replication origin | This study |
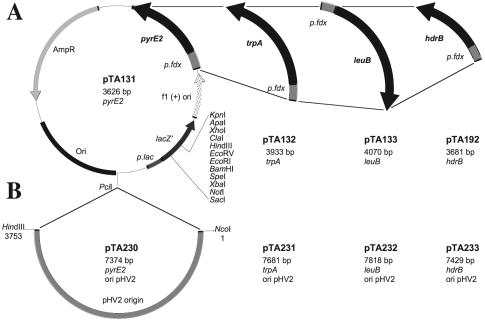
FIG. 5.
Plasmid vectors marked with pyrE2, leuB, trpA, and hdrB. (A) Gene knockout plasmids. The pyrE2, leuB, trpA, and hdrB genes were placed under control of the ferredoxin promoter and cloned in pBluescript II to generate pTA131, pTA132, pTA133, and pTA192, respectively. (B) Shuttle vectors. The pHV2 replication origin from pWL102 (7, 22) was cloned in pTA131, pTA132, pTA133, and pTA192 to generate pTA230, pTA231, pTA232, and pTA233, respectively. Some sites in the polylinker are not unique in the shuttle vectors; full plasmid maps are available on request.
(i) Integrative plasmids.
To generate pTA131, a _Bam_HI-_Xba_I fragment from pGB70 (3) containing the pyrE2 gene under control of the constitutive ferredoxin promoter was inserted into the _Psi_I site of pBluescript II. The _Psi_I site lies outside the polylinker in the lacZ promoter; therefore, the blue-white screening capability is retained. To generate pTA133, pTA132, and pTA192, the coding regions of leuB, trpA, and hdrB were amplified from pTA44, pTA49, and pD4 (26), respectively. The primers used (Table 2) incorporated _Pci_I or _Bsp_HI sites at the ATG start codon, which were compatible with the _Nco_I site at the 3′ end of the ferredoxin promoter. The ensuing fusion constructs were inserted into the _Psi_I site of pBluescript II.
(ii) Shuttle vectors.
An _Nco_I-_Hin_dIII fragment of pWL102 containing the pHV2 replication origin (7, 22) was inserted into the _Pci_I sites of pTA131, pTA132, pTA133, and pTA192 to generate pTA230, pTA231, pTA232, and pTA233, respectively.
DNA sequence analysis.
DNA database searches of the H. volcanii genome were performed by using NCBI BLAST for Mac OS X (ftp://ftp.ncbi.nih.gov/blast/executables/), and sequence files were downloaded from the University of Scranton (http://wit-scranton.mbi.scranton.edu/Haloferax/). Genomic clones were sequenced by using an in-house service and were used to amend the University of Scranton data. Sequences of pyrE2 (3), hdrB (26), and the trpCBA operon (20) have been published previously.
Nucleotide sequence accession number.
The corrected nucleotide sequence of the H. volcanii leuCDB operon has been deposited in the EMBL nucleotide sequence database under accession number AJ571689.
RESULTS
Construction of H. volcanii DS70 Δ_pyrE2_ strain.
A Δ_pyrE2_ mutant was constructed previously in the H. volcanii WFD11 background (3). Since WFD11 strains (22) have been reported to suffer from growth reduction and plasmid instability (31), we implemented the pyrE2 gene knockout system in the improved H. volcanii strain DS70 (31) using the method described by Bitan-Banin et al. (3). DS70 cells were transformed to novobiocin resistance with pGB68, which contains the 850-bp upstream and 850-bp downstream flanking sequences of pyrE2 (3). Transformants (pop-in) were screened by Southern blot hybridization, and a clone (H18) was selected which had integrated at the pyrE2 locus and not gyrB; since novobiocin resistance is encoded by a mutant allele of the DNA gyrase gene gyrB (15), plasmids containing this marker can integrate at the chromosomal gyrB locus (which was the case in two of the five transformants examined). Excision of pGB68 (pop-out) was performed by propagating H18 for ∼30 generations in rich medium (Hv-YPC) in the absence of novobiocin and plating on Casamino Acids (Hv-Ca) agar containing either uracil alone or uracil and 5-FOA. Approximately 2% of the cells were 5-FOA resistant and were subsequently determined to be auxotrophic for uracil and sensitive to novobiocin. These clones were analyzed by Southern blot hybridization, and a strain in which pGB68 had been excised, resulting in deletion of pyrE2, was designated H26.
Cloning of leuB and flanking sequences.
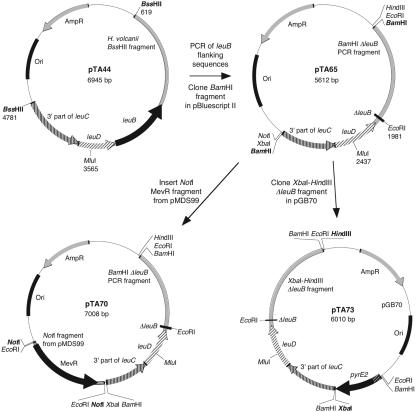
An operon containing the leucine biosynthesis genes leuB, leuC, and leuD was identified in the H. volcanii genome sequence. The leuB gene, encoding 3-isopropylmalate dehydrogenase, was chosen for development as a genetic marker, since it is the terminal gene in the operon and a deletion would not have a polar effect on transcription of leuC and leuD. To clone the gene and its flanking sequences, a fragment of leuB was amplified by PCR and used to probe a Southern blot of H. volcanii chromosomal DNA digested with _Bss_HII. A 4.2-kb DNA fragment was found to hybridize with the probe. A genomic DNA mini-library of 4.2-kb _Bss_HII fragments was constructed in pBluescript II and screened by colony hybridization by using the leuB fragment probe. A clone (pTA44) was sequenced and found to contain the 972-bp leuB gene, as well as 1,604 bp of upstream flanking sequences and 1,584 bp of downstream flanking sequences (Fig. 2).
FIG. 2.
Construction of leuB deletion plasmids. The genomic leuB clone pTA44 contains a 4,162-bp genomic DNA Bss_HII fragment cloned in pBluescript II. Sequences flanking leuB were amplified from pTA44 and cloned in pBluescript II to generate the Δ_leuB construct pTA65. The mevinolin resistance fragment from pMDS99 (31) was inserted into pTA65 to generate pTA70. Alternatively, the Δ_leuB_ construct from pTA65 was cloned in the _pyrE2_-marked gene knockout plasmid pGB70 (3), generating pTA73. Only relevant sites are shown; full plasmid maps are available on request.
Deletion of leuB and phenotypic analysis.
To delete leuB, a 1,268-bp fragment upstream of leuB and a 1,395-bp downstream fragment were amplified by PCR by using pTA44 as a template. The internal primers contained _Eco_RI sites used to ligate the PCR products, and the external primers contained Bam_HI sites used to clone the Δ_leuB fragment in pBluescript II SK+, generating pTA65 (Fig. 2). A _Not_I fragment from pMDS99 (31) containing the mevinolin resistance gene (MevR) was inserted at the _Not_I site of pTA65 to generate pTA70. Alternatively, an _Xba_I-Hin_dIII fragment of pTA65 containing the Δ_leuB construct was inserted at the _Xba_I and _Hin_dIII sites of the _pyrE2_-marked plasmid pGB70 (3), generating pTA73 (Fig. 2).
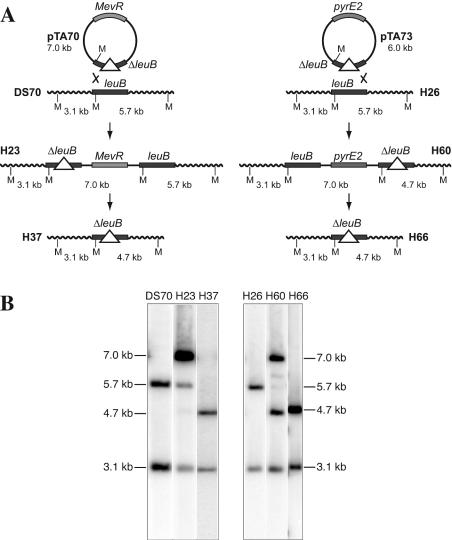
H. volcanii DS70 was transformed to mevinolin resistance with pTA70, and transformants were screened for integration at the leuB locus by Southern blot hybridization (Fig. 3); this was observed in all eight transformants analyzed. One clone was chosen (H23), and excision of pTA70 was performed by propagating H23 in the absence of mevinolin. Colonies were screened by replica plating on rich agar with and without mevinolin, as well as minimal agar (Hv-Min). Mevinolin-sensitive clones that failed to grow on minimal agar without added leucine were analyzed by Southern blot hybridization. A strain in which pTA70 had been excised, resulting in deletion of 942 bp, including leuB, was designated H37 (Fig. 3).
FIG. 3.
Deletion of leuB gene. (A) Plasmids pTA70 and pTA73 were constructed as described in the legend to Fig. 2. Integration of pTA70 into the chromosome of DS70 by homologous recombination upstream of leuB resulted in strain H23. Loss of the plasmid by intrachromosomal recombination (Fig. 1) resulted in the Δ_leuB_ strain H37. Integration of pTA73 (in H26) by recombination downstream of leuB yielded strain H60, and loss of the plasmid resulted in the Δ_leuB_ strain H66. Integration and deletion events were monitored by digestion with Mlu_I (M), resulting in the fragments indicated. (B) Southern blot analysis of Δ_leuB strain construction. Genomic DNA were prepared from the strains indicated, digested with _Mlu_I, and probed with the flanking regions of leuB.
To construct Δ_pyrE2_ Δ_leuB_ strains, the H. volcanii DS70 Δ_pyrE2_ strain H26 was transformed to uracil prototrophy with pTA73, and transformants were screened for integration at the leuB locus (Fig. 3). One clone was chosen (H60), and excision of pTA73 was performed by propagating H60 in rich medium and plating the culture on Casamino Acids (Hv-Ca) agar containing either uracil alone or uracil and 5-FOA. Approximately 2% of the cells were 5-FOA resistant (Ura−) and were screened by replica plating on minimal agar with and without added leucine. Five of 30 Ura− clones tested were auxotrophic for leucine and were analyzed by Southern blotting (Fig. 3B). A strain in which pTA73 had been excised, resulting in deletion of leuB, was designated H66. A Δ_pyrE2_ Δ_leuB_ strain was made in a similar manner in the WFD11 background by transforming WR480 (3) with pTA73. The pop-in strain was designated H30, and the Δ_leuB_ pop-out strain was designated H40.
Cloning of trpA and flanking sequences.
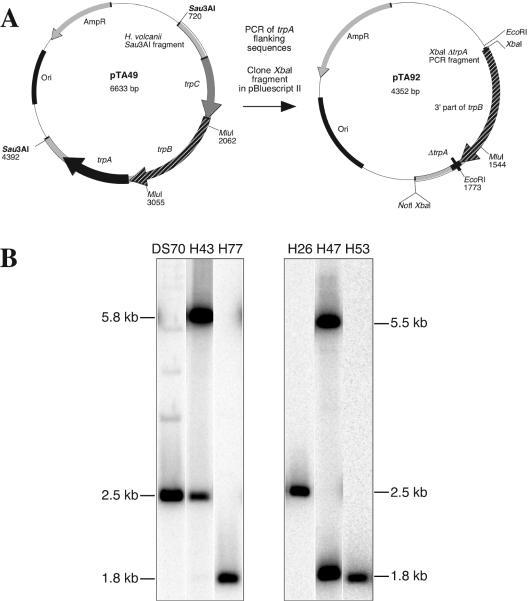
To clone the operon containing the tryptophan biosynthesis genes trpC, trpB, and trpA and their flanking sequences (20), a fragment of trpA was amplified and used to probe a Southern blot of H. volcanii DNA digested with _Sau_3AI. As predicted (20), a 3.7-kb DNA fragment hybridized with the probe. A genomic DNA mini-library of 3.7-kb _Sau_3AI fragments was constructed in pBluescript II and screened by colony hybridization. A clone (pTA49) (Fig. 4A) was isolated that contained the 834-bp trpA gene, as well as 2,531 bp of upstream flanking sequences and 311 bp of downstream flanking sequences. The trpA gene, encoding the 277-amino-acid tryptophan synthase alpha subunit, was chosen for development as a genetic marker since it is the terminal gene in the operon.
FIG. 4.
Deletion of trpA gene. (A) Construction of trpA deletion plasmids. The genomic trpA clone pTA49 contains a 3,676-bp genomic DNA Sau_3AI fragment cloned in pBluescript II. Sequences flanking trpA were amplified from pTA49 and cloned in pBluescript II to generate the Δ_trpA plasmid pTA92. The mevinolin resistance fragment from pMDS99 (31) was inserted into pTA92 to generate pTA93 (data not shown; similar to pTA70 in Fig. 2). Alternatively, the Δ_trpA_ construct from pTA92 was cloned in pGB70 (3), generating pTA95 (data not shown; similar to pTA73 in Fig. 2). Plasmid maps are available on request. (B) Deletion of the trpA gene in strains H77 and H53 was analyzed by _Mlu_I digestion and Southern blot hybridization by using flanking regions of trpA as a probe (similar to the leuB deletion in Fig. 3). Integration of pTA93 into the chromosome of DS70 by recombination upstream of trpA gave a novel 5.8-kb _Mlu_I fragment, producing strain H43. Loss of the plasmid, which deleted trpA (1.8-kb _Mlu_I fragment instead of 2.5-kb Mlu_I fragment) resulted in Δ_trpA strain H77. Integration of pTA95 into H26 by recombination downstream of trpA gave a novel 5.5-kb Mlu_I fragment, producing strain H47, and loss of the plasmid resulted in Δ_trpA strain H53.
Deletion of trpA and phenotypic analysis.
To delete trpA, a 1,048-bp fragment upstream of trpA and a 355-bp downstream fragment were amplified by PCR by using pTA49. The internal primers contained _Eco_RI sites used to ligate the PCR products, and the external primers contained Xba_I sites used to clone the Δ_trpA fragment in pBluescript II, generating pTA92 (Fig. 4A). The _Not_I MevR fragment from pMDS99 (31) was inserted at the _Not_I site of pTA92 to generate pTA93 (data not shown; similar to pTA70 in Fig. 2). Alternatively, an Xba_I fragment of pTA92 containing the Δ_trpA construct was inserted at the _Xba_I site of pGB70 (3), generating pTA95 (data not shown; similar to pTA73 in Fig. 2).
Plasmid pTA93 was used to transform H. volcanii DS70. Transformants were screened for integration at trpA by Southern blotting (Fig. 4B). One integrant was chosen (H43), and excision of pTA93 was performed as described above. Colonies were also screened by replica plating on Casamino Acids (Hv-Ca) agar, which contained no detectable tryptophan. Mevinolin-sensitive clones exhibiting tryptophan auxotrophy on Casamino Acids agar were analyzed by Southern blotting. A strain in which pTA93 had been excised to obtain a 744-bp deletion of trpA was designated H77 (Fig. 4). A Δ_trpA_ strain was made in a similar manner in the WFD11 background, by transforming WR340 (3) with pTA93. The pop-in strain was designated H42, and the Δ_trpA_ pop-out strain was designated H76.
To construct a Δ_pyrE2_ Δ_trpA_ strain, H. volcanii H26 was transformed with pTA95, and transformants were screened for integration at trpA. One integrant was chosen (H47) (Fig. 4B), and excision of pTA95 was performed as described above. 5-FOA-resistant (Ura−) cells were screened for tryptophan auxotrophy by replica plating on Casamino Acids agar and were analyzed by Southern blot hybridization (Fig. 4B). A strain in which pTA95 had been excised to delete trpA was designated H53. A Δ_pyrE2_ Δ_trpA_ strain was made in a similar manner in the WFD11 background, by transforming WR480 (3) with pTA95. The pop-in strain was designated H45, and the Δ_trpA_ pop-out strain was designated H52.
We examined the potential of Δ_trpA_ strains in a counterselectable system similar to that based on pyrE2. In S. cerevisiae, 5-fluoroanthranilic acid has been used for counterselection of tryptophan biosynthesis genes (30). This compound is converted to toxic 5-fluorotryptophan in trp+ cells, which is analogous to the action on 5-FOA in uracil biosynthesis (4). We tested a number of anthranilic acid derivatives, including 4-, 5-, and 6-fluoroanthranilic acids and 5- and 6-methylanthranilic acids, as well as 5-fluoroindole, for the ability to select against trpA+ strains but not Δ_trpA_ strains. Strains H26 and H53 were tested by plating on Casamino Acids agar containing between 0.05 and 1 mg of the anthranilic acid derivatives per ml; tryptophan was added at a relative concentration of 10 to 40% to support growth of Δ_trpA_ cells. None of these compounds discriminated between H26 (trpA+) and H53 (Δ_trpA_) cells; the anthranilic acid derivatives prevented growth of both strains at concentrations above 0.25 mg/ml, and 5-fluoroindole was toxic at all concentrations tested. This was most likely due to feedback inhibition of tryptophan biosynthesis, leading to insufficient conversion of anthranilic acid derivatives to toxic fluoro- or methyltryptophan, and a failure to discriminate between trpA+ and Δ_trpA_ strains. In addition, the anthranilic acid derivatives most probably exhibited nonspecific toxicity at higher concentrations, affecting both trpA+ and Δ_trpA_ cells.
Construction of strains with deletions in pyrE2, leuB, trpA, and hdrB.
The hdrB marker is a useful addition to the current genetic repertoire, as deletion of this gene confers thymidine auxotrophy in rich medium (Hv-YPC) (26). We therefore constructed a Δ_hdrB_ mutant in the H. volcanii DS70 Δ_pyrE2_ strain H26. Sequences flanking hdrB (577 bp upstream and 292 bp downstream) were amplified by PCR by using the genomic clone pD4 as a template (26) and were cloned in the pyrE2_-marked gene knockout plasmid pTA131 (see below) (Fig. 5A) to generate the Δ_hdrB construct pTA155. H. volcanii H26 was transformed with pTA155, and transformants were screened for integration at hdrB (data not shown). One integrant was chosen (H90), and excision of pTA155 was performed as described above. 5-FOA-resistant (Ura−) cells were screened for thymidine auxotrophy by replica plating on rich agar. A strain in which pTA155 had been excised to delete hdrB was designated H98.
We constructed strains with combinations of the deletions described above. A Δ_pyrE2_ Δ_leuB_ Δ_trpA_ strain was made by transforming H53 with pTA73; the pop-in strain was designated H111, and the Δ_leuB_ pop-out strain was designated H119. Derivatives of H53, H66, and H119 with an additional deletion of hdrB were constructed by transforming these strains with pTA155; the pop-in strains were designated H91, H92, and H126, respectively, and the Δ_hdrB_ pop-out strains were designated H99, H100, and H133, respectively. All of the strains described here are listed in Table 1.
Construction of plasmid vectors by using pyrE2, leuB, trpA, and hdrB as selectable markers.
To construct an improved _pyrE2_-marked gene knockout plasmid, a fragment from pGB70 containing the pyrE2 gene under control of the constitutive ferredoxin promoter of Halobacterium salinarum (27) was inserted into pBluescript II. This plasmid (pTA131) (Fig. 5A) retained the blue-white screening facility of pBluescript. Similar plasmids were made by using leuB, trpA, and hdrB as selectable markers. The coding regions of these genes were amplified from pTA44, pTA49, and pD4, fused to the ferredoxin promoter, and inserted into pBluescript II to generate pTA133, pTA132, and pTA192, respectively (Fig. 5A).
Shuttle vectors were derived from these plasmids by inserting the replication origin of the H. volcanii episome pHV2 (Fig. 5B); both strain WFD11 and strain DS70 have been cured of this indigenous plasmid (7, 22, 31). The shuttle vectors were able to transform the corresponding H. volcanii deletion strains to prototrophy for the appropriate markers. The transformation efficiencies were ∼105 CFU/μg of DNA, as expected. Dual-resistance vectors based on pMDS20 (14), which also contained the novobiocin resistance gene, were also constructed (details are available on request).
Construction of a Δ_lhr_ strain by using the Δ_pyrE2_ Δ_trpA_ strain.
In the pop-in-pop-out gene knockout system, spontaneous excision of the integrated plasmid can occur in one of two ways, either restoring the gene to wild-type information or resulting in a deletion (Fig. 1A). Even if both outcomes are equally likely, the mutant rapidly becomes underrepresented (in the population of 5-FOA-resistant cells) if the gene deletion leads to a slow-growth phenotype. By replacing the gene to be deleted with the trpA marker and plating the culture on Casamino Acids medium with 5-FOA (but no added tryptophan), it is possible to select directly for pop-out events that lead to the mutation (Fig. 1B).
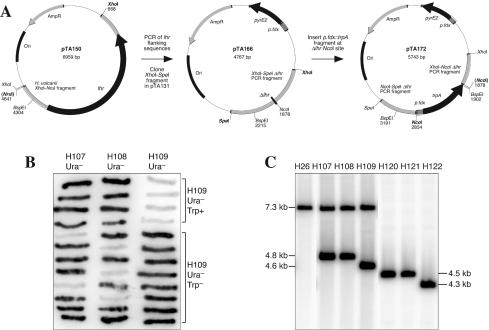
To demonstrate that the trpA pyrE2 system allows direct selection for gene knockouts that might otherwise be difficult to recover, we deleted the lhr gene, which was identified in the genome sequence on the basis of the homology of its product to the Lhr protein of E. coli. Lhr is a member of helicase superfamily II (28) and is well conserved among prokaryotes, although the C-terminal ∼650 amino acids of the E. coli protein are found only in bacteria. A 3,974-bp _Xho_I-_Nru_I fragment of H. volcanii genomic DNA, containing the 2,757-bp lhr gene, 677 bp of upstream flanking sequences, and 540 bp of downstream flanking sequences, was cloned in pBluescript II to generate pTA150 (Fig. 6A). To delete lhr, a 653-bp upstream fragment and a 551-bp downstream fragment were amplified by PCR by using pTA150. The internal primers contained _Nco_I sites used to ligate the PCR products, and the external primers contained _Xho_I and Spe_I sites used to clone the Δ_lhr fragment in pTA131, generating pTA166 (Fig. 6A). A 972-bp fragment containing the trpA gene under control of the ferredoxin promoter (see pTA132 in Fig. 5A) was inserted into the _Nco_I site at the lhr deletion to generate pTA172 (Fig. 6A).
FIG. 6.
Deletion of lhr gene. (A) Construction of lhr deletion plasmid. The genomic lhr clone pTA150 was used to amplify the flanking regions of lhr, which were inserted into pTA131 to generate the Δ_lhr_ construct pTA166. A fragment containing the trpA gene under control of the ferredoxin promoter (p.fdx) was inserted at the site of the lhr deletion (Nco_I), generating the Δ_lhr::trpA plasmid pTA172. (B) Twelve 5-FOA-resistant (Ura−) derivatives of pop-in strains H107 to H109 were grown on rich agar, transferred to a nylon filter, and probed with the lhr coding sequence. In the case of H109, the four Ura− clones with lhr deleted had previously been shown to be prototrophic for tryptophan (Trp+), whereas the remaining eight clones in which lhr was not deleted were Trp−. (C) Deletion of the lhr gene was analyzed by _Bsp_EI digestion and Southern blot hybridization by using the downstream flanking region of lhr as a probe. Integration of pTA166 into the chromosomes of H26 and H53 gave a novel 4.8-kb _Bsp_EI fragment, producing strains H107 and H108, respectively, and loss of pTA166, which deleted lhr (4.5-kb _Bsp_EI fragment instead of 7.3-kb Bsp_EI fragment), resulted in Δ_lhr strains H120 and H121, respectively. Integration of pTA172 in the chromosome of H53 gave a novel 4.6-kb _Bsp_EI fragment, producing strain H109, and loss of pTA172, which deleted lhr (4.3-kb Bsp_EI fragment), resulted in Δ_lhr::trpA strain H122.
H26 was transformed with pTA166, and H53 was transformed with pTA166 or pTA172. Integrants at lhr were verified (H107 to H109, respectively) (Fig. 6C), and excision of pTA166 and pTA172 was performed as usual. Cultures were plated on Casamino Acids agar containing uracil and 5-FOA with or without added tryptophan. Approximately 2% of the cells were 5-FOA resistant (Ura−), and in the case of H109 (transformed with Δ_lhr_::trpA plasmid pTA172), 4 of the 30 Ura− clones tested were prototrophic for tryptophan. Ura− clones were screened for deletion of lhr by colony hybridization by using the lhr coding sequence as a probe (Fig. 6B). All four of the Trp+ derivatives of H109 were Δ_lhr_::trpA, while in the remaining Trp− derivatives lhr was not deleted. Among derivatives of H107 and H108 (transformed with Δ_lhr_ plasmid pTA172) only 2 of 12 and 3 of 12 of the Ura− clones, respectively, proved to be Δ_lhr_ (Fig. 6B). Deletions were confirmed by Southern blot hybridization (Fig. 6C), and Δ_lhr_ derivatives of H107 to H109 were designated H120 to H122, respectively. Δ_lhr_ mutants did not show any detectable growth deficiency in rich or minimal medium and were no more sensitive to UV or γ radiation than isogenic lhr+ strains were.
DISCUSSION
A cornerstone of genetic analysis is the ability to manipulate the genome. Traditionally, this has been done by chemical mutagenesis, followed by laborious screening for the appropriate phenotype. In the postgenomic era, when homologues are readily identifiable in model organisms, it is more expedient to perform targeted gene knockout and characterize the phenotype of the mutant. Selectable markers provide the means to accomplish this end.
The development of antibiotic resistance markers for the archaea has been hampered by the lack of drug targets. Bacterial antibiotics are safe for medical use as their targets are generally not found in eukaryotic cells. Due to the greater similarity of the archaea to eukaryotes, it is hardly surprising that most commonly used antibiotics are ineffective against archaea (13). The few antibiotics currently available for H. volcanii have shortcomings. For example, the mevinolin resistance marker is an up-promoter mutation of the chromosomal gene hmgA (21). Recombination between the chromosomal gene and the resistance marker can therefore lead to constitutive antibiotic resistance. This problem was recently alleviated by development of a mevinolin resistance marker from Haloarcula hispanica, which is stably maintained in H. volcanii (31). However, spontaneous resistance to mevinolin can still arise (at an inconveniently high frequency) by promoter point mutation or amplification of the hmg gene (T.A., unpublished observations). Stable auxotrophic mutants provide a solution to these problems.
We have developed leuB and trpA as selectable markers, in conjunction with existing systems based on pyrE2 and hdrB, since this suite of genes takes full advantage of the media available for H. volcanii (Table 4). In order to ensure that the mutants are stable, complete gene deletions were constructed. The coding sequence of the deleted gene can then be used as a selectable marker on a replicative (shuttle) vector without the risk of integration by homologous recombination. We have generated shuttle vectors and integrative plasmids for gene knockouts that complement these deletions (Fig. 5). Implementation of the leuB, trpA, and hdrB deletions in the Δ_pyrE2_ background provides the widest variety of genetic markers available in any archaeal species.
TABLE 4.
Growth characteristics of selected H. volcanii mutants
| Strain | Genotype | Growth on: | ||
|---|---|---|---|---|
| Hv-YPC | Hv-Ca | Hv-Min | ||
| H26 | Δ_pyrE2_ | + | Ura− | Ura− |
| H37 | Δ_leuB_ | + | + | Leu− |
| H77 | Δ_trpA_ | + | Trp− | Trp− |
| H66 | Δ_pyrE2_ Δ_leuB_ | + | Ura− | Ura− Leu− |
| H53 | Δ_pyrE2_ Δ_trpA_ | + | Ura− Trp− | Ura− Trp− |
| H98a | Δ_pyrE2_ Δ_hdrB_ | Thy− | Ura− Thy− | Ura− Thy− |
The genetic tools described here, particularly the trpA pyrE2 system, should be useful for isolation of mutants that are deleterious and therefore difficult to recover. By using a construct in which the gene of interest is replaced with the trpA marker, it is possible to select directly for deletion events (Fig. 1B). We demonstrated the utility of this system with a deletion of the lhr gene of H. volcanii (Fig. 6). Complete failure to recover 5-FOA-resistant Trp+ cells from such a pop-in strain would be a strong indication that the gene deletion is lethal. This facility should in turn permit development of more sophisticated genetic tools, such as synthetic lethal screening methods (11).
Acknowledgments
We thank Steven Marsden for help with the leuB and trpA deletions, Gili Bitan-Banin for valuable advice, and Ed Bolt for critical reading of the manuscript.
This work was supported by a Wellcome Trust project grant to T.A. and R.G.L. and by a Royal Society University Research Fellowship to T.A.
REFERENCES
- 1.Beggs, J. D. 1978. Transformation of yeast by a replicating hybrid plasmid. Nature 275**:**104-109. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. D., and S. P. Jackson. 2001. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 4**:**208-213. [DOI] [PubMed] [Google Scholar]
- 3.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185**:**772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197**:**345-346. [DOI] [PubMed] [Google Scholar]
- 5.Brown, J. R., and W. F. Doolittle. 1997. Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev. 61**:**456-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273**:**1058-1073. [DOI] [PubMed] [Google Scholar]
- 7.Charlebois, R. L., W. L. Lam, S. W. Cline, and W. F. Doolittle. 1987. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc. Natl. Acad. Sci. 84**:**8530-8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cline, S. W., W. L. Lam, R. L. Charlebois, L. C. Schalkwyk, and W. F. Doolittle. 1989. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35**:**148-152. [DOI] [PubMed] [Google Scholar]
- 9.DeLong, E. F., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65**:**5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis, P. P. 1997. Ancient ciphers: translation in Archaea. Cell 89**:**1007-1010. [DOI] [PubMed] [Google Scholar]
- 11.Forsburg, S. L. 2001. The art and design of genetic screens: yeast. Nat. Rev. Genet. 2**:**659-668. [DOI] [PubMed] [Google Scholar]
- 12.Graham, D. E., R. Overbeek, G. J. Olsen, and C. R. Woese. 2000. An archaeal genomic signature. Proc. Natl. Acad. Sci. 97**:**3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilpert, R., J. Winter, W. Hammes, and O. Kandler. 1981. The sensitivity of archaebacteria to antibiotics. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. Abt. 1 Orig. Reihe C 2**:**11-20. [Google Scholar]
- 14.Holmes, M., F. Pfeifer, and M. Dyall-Smith. 1994. Improved shuttle vectors for Haloferax volcanii including a dual-resistance plasmid. Gene 146**:**117-121. [DOI] [PubMed] [Google Scholar]
- 15.Holmes, M. L., and M. L. Dyall-Smith. 1991. Mutations in DNA gyrase result in novobiocin resistance in halophilic archaebacteria. J. Bacteriol. 173**:**642-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes, M. L., and M. L. Dyall-Smith. 1990. A plasmid vector with a selectable marker for halophilic archaebacteria. J. Bacteriol. 172**:**756-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes, M. L., and M. L. Dyall-Smith. 2000. Sequence and expression of a halobacterial beta-galactosidase gene. Mol. Microbiol. 36**:**114-122. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, M. L., S. D. Nuttall, and M. L. Dyall-Smith. 1991. Construction and use of halobacterial shuttle vectors and further studies on Haloferax DNA gyrase. J. Bacteriol. 173**:**3807-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komori, K., S. Sakae, H. Shinagawa, K. Morikawa, and Y. Ishino. 1999. A Holliday junction resolvase from Pyrococcus furiosus: functional similarity to Escherichia coli RuvC provides evidence for conserved mechanism of homologous recombination in Bacteria, Eukarya, and Archaea. Proc. Natl. Acad. Sci. 96**:**8873-8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam, W. L., A. Cohen, D. Tsouluhas, and W. F. Doolittle. 1990. Genes for tryptophan biosynthesis in the archaebacterium Haloferax volcanii. Proc. Natl. Acad. Sci. 87**:**6614-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam, W. L., and W. F. Doolittle. 1992. Mevinolin-resistant mutations identify a promoter and the gene for a eukaryote-like 3-hydroxy-3-methylglutaryl-coenzyme A reductase in the archaebacterium Haloferax volcanii. J. Biol. Chem. 267**:**5829-5834. [PubMed] [Google Scholar]
- 22.Lam, W. L., and W. F. Doolittle. 1989. Shuttle vectors for the archaebacterium Halobacterium volcanii. Proc. Natl. Acad. Sci. 86**:**5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacNeill, S. A. 2001. Understanding the enzymology of archaeal DNA replication: progress in form and function. Mol. Microbiol. 40**:**520-529. [DOI] [PubMed] [Google Scholar]
- 24.Mevarech, M., and R. Werczberger. 1985. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 162**:**461-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullakhanbhai, M. F., and H. Larsen. 1975. Halobacterium volcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch. Microbiol. 104**:**207-214. [DOI] [PubMed] [Google Scholar]
- 26.Ortenberg, R., O. Rozenblatt-Rosen, and M. Mevarech. 2000. The extremely halophilic archaeon Haloferax volcanii has two very different dihydrofolate reductases. Mol. Microbiol. 35**:**1493-1505. [DOI] [PubMed] [Google Scholar]
- 27.Pfeifer, F., J. Griffig, and D. Oesterhelt. 1993. The fdx gene encoding the [2Fe-2S] ferredoxin of Halobacterium salinarium (H. halobium). Mol. Gen. Genet. 239**:**66-71. [DOI] [PubMed] [Google Scholar]
- 28.Reuven, N. B., E. V. Koonin, K. E. Rudd, and M. P. Deutscher. 1995. The gene for the longest known Escherichia coli protein is a member of helicase superfamily II. J. Bacteriol. 177**:**5393-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Toyn, J. H., P. L. Gunyuzlu, W. H. White, L. A. Thompson, and G. F. Hollis. 2000. A counterselection for the tryptophan pathway in yeast: 5-fluoroanthranilic acid resistance. Yeast 16**:**553-560. [DOI] [PubMed] [Google Scholar]
- 31.Wendoloski, D., C. Ferrer, and M. L. Dyall-Smith. 2001. A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2. Microbiology 147**:**959-964. [DOI] [PubMed] [Google Scholar]
- 32.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. 87**:**4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]