Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome (original) (raw)
. Author manuscript; available in PMC: 2013 Oct 12.
SUMMARY
Pathogenic simian immunodeficiency virus (SIV) infection is associated with enteropathy which likely contributes to AIDS progression. To identify candidate etiologies for AIDS enteropathy, we used next generation sequencing to define the enteric virome during SIV infection in nonhuman primates. Pathogenic, but not non-pathogenic, SIV infection was associated with significant expansion of the enteric virome. We identified at least 32 previously undescribed enteric viruses during pathogenic SIV infection and confirmed their presence using viral culture and PCR testing. We detected unsuspected mucosal adenovirus infection associated with enteritis as well as parvovirus viremia in animals with advanced AIDS, indicating the pathogenic potential of SIV-associated expansion of the enteric virome. No association between pathogenic SIV infection and the family-level taxonomy of enteric bacteria was detected. Thus, enteric viral infections may contribute to AIDS enteropathy and disease progression. These findings underline the importance of metagenomic analysis of the virome for understanding AIDS pathogenesis.
INTRODUCTION
HIV infection of humans and pathogenic SIV infection of rhesus monkeys causes progressive immunocompromise and acquired immune deficiency syndrome (AIDS). The rate of progression to AIDS correlates with loss of CD4 T cells, lentivirus RNA levels in the blood, and systemic immune activation (Brenchley and Douek, 2012; Brenchley et al., 2006b; Sandler and Douek, 2012). Thus, lentivirus-infected humans and primates that progress to AIDS exhibit markers of systemic immune activation including elevated serum and tissue cytokines such as type I interferon, increased serum soluble CD14 and LPS-binding protein (LBP), and alterations in T cell activation markers. Systemic immune activation is, in turn, associated with damage to the intestinal epithelium and translocation of as-yet-undefined immunostimulatory pathogen-associated molecular patterns (PAMPS) or antigens into tissues and the blood (Brenchley and Douek, 2012; Brenchley et al., 2006b; Estes et al., 2010; Sandler and Douek, 2012).
Systemic immune activation in SIV-infected rhesus monkeys is associated with breakdown of the intestinal epithelial lining (Estes et al., 2010; Sandler and Douek, 2012). Interestingly, natural hosts for SIV such as African green monkeys develop persistent high level viremia but do not develop AIDS (termed herein ‘non-pathogenic’ SIV infection) (Brenchley and Douek, 2012; Brenchley et al., 2010; Sodora et al., 2009). Further, these animals do not exhibit systemic immune activation or translocation of intestinal PAMPS into the circulation (Brenchley and Douek, 2012; Brenchley et al., 2010; Pandrea et al., 2008; Sodora et al., 2009). However, when LPS is administered to non-pathogenically SIV-infected African green monkeys, systemic immune activation and increased SIV replication are observed (Pandrea et al., 2008). This suggests a feed-forward mechanism contributing to AIDS progression in which intestinal epithelial damage leads to translocation of PAMPs or antigens into tissues which contributes to systemic immune activation, increased lentivirus replication, progressive immune deficiency and AIDS (Brenchley and Douek, 2012; Brenchley et al., 2006a; Brenchley et al., 2006b; Sandler and Douek, 2012).
Despite the importance of intestinal barrier damage to AIDS progression, the mechanisms responsible for AIDS enteropathy are not understood. One obvious possibility is that immunodeficiency leads to epithelial damage by intestinal viruses or other pathogens. The mammalian virome and bacterial microbiome is extremely complex and can contribute to immune status and disease in a range of settings (Barton et al., 2007; Costello et al., 2009; Kau et al., 2011; Virgin and Todd, 2011; Virgin et al., 2009). A prior study that utilized 16S rDNA sequencing, which was unable to detect viruses, found no discernible differences in the diversity of bacteria associated with SIV infection (McKenna et al., 2008). The virome is a subset of the metagenome that may be defined to include both viruses that infect eukaryotic cells and phages that infect other members of the microbiome. Herein, we will define viruses that infect eukaryotic cells as the virome (Virgin et al., 2009). Indeed, primate species used in SIV research can be infected with a range of enteropathogenic viruses (Farkas et al., 2008; Oberste et al., 2002, 2007; Sasseville and Mansfield, 2010; Wang et al., 2007).
We hypothesized that current diagnostic approaches miss potential viral causes for epithelial damage during SIV infection, and used next generation sequencing (NGS) to define the enteric virome during SIV infection. We observed that pathogenic SIV infection in rhesus monkeys, but not non-pathogenic SIV infection of African green monkeys was associated with a substantial expansion of the enteric virome, and using very conservative criteria identified at least 32 previously undescribed viruses from multiple pathogenic viral genera. In particular, adenoviruses detected by NGS during pathogenic SIV infection caused unexpected enteritis, indicating that infection with these viruses can be associated with pathology seen in AIDS enteropathy. Further, enteric parvoviruses detected in feces were found in the circulation during advanced AIDs. However, we did not detect SIV-associated changes in the composition of the bacterial microbiome. We speculate that the enteric virome contributes to the progression of SIV infection to AIDS by fostering intestinal epithelial damage and systemic immune activation via release of pathogens as well as bacterial, viral, fungal or other PAMPs and antigens into host tissues and the systemic circulation. This study highlights the use of shotgun sequencing to detect a broad range of viruses present during pathogenic SIV infection.
RESULTS
Defining the enteric virome of SIV-infected and control monkeys
To define the effects of pathogenic and non-pathogenic SIV infection on the enteric virome, we shotgun sequenced libraries of fecal RNA + DNA from four independent cohorts of monkeys, each comprising SIV infected and uninfected control animals. Pathogenically SIV-infected rhesus monkeys were housed at the New England Primate Research Center (NEPRC, sampled at 24 and 64 weeks post SIV infection) or the Tulane National Primate Research Center (TNPRC) (Table 1). Analysis of this cohort confirmed SIV viremia in infected animals and revealed the expected decreases in CD4 T cell counts and increases in serum LBP levels consistent with intestinal leakage and consequent systemic immune activation at both 24 and 64 weeks after infection (Figure S1). As expected, the set point level of SIV in the serum correlated with rapid progression to AIDS and death (not shown). Non-pathogenically SIV-infected African green monkeys were housed at the National Institutes of Health (NIH, vervet monkeys) or the NEPRC (sabaeus monkeys).
Table 1.
see also Figure S1. Cohorts and sequences analyzed
| Animal Cohort | Type of monkey | Animal numbers | Total sequences (average length) | Unique sequences (average length) | Total sequences per sample | Unique sequences per sample | |
|---|---|---|---|---|---|---|---|
| NEPRC1 (24 wpi2) | Rhesus | 22 Control | 22 SIV+ | 899,947 (358 bp) | 356,521 (357 bp) | 4,689–51,870 | 594–26,838 |
| NEPRC (64 wpi) | Rhesus | 22 Control | 12 SIV+ | 705,429 (341 bp) | 263,430 (345 bp) | 6,132–59,847 | 1,080–33,982 |
| TNPRC3 | Rhesus | 29 Control | 13 SIV+ | 1,409,046 (296 bp) | 557,518 (294 bp) | 9,188–8,9974 | 3,666–3,3613 |
| NIH4 | African green | 19 Control | 19 SIV+ | 1,382,171 (300 bp) | 425,524 (301 bp) | 3,259–127,567 | 1,382–33,464 |
| NEPRC | African green | 6 Control | 10 SIV + | 612,612 (293 bp) | 187,807 (279 bp) | 8,287–194,880 | 2,118–55,158 |
Total RNA + DNA from fecal material were sequenced using 454 technology to leverage the resulting long sequences for robust assessment of taxonomy and assembly of viral genomes (Table 1). There was no statistical correlation between SIV infection and either the number of total or unique sequences (viral plus other) obtained within any of the four cohorts. For each cohort, sequences were analyzed using two approaches. In the first, the taxonomic structure of the sequences was analyzed using MEGAN version 4.62.3 [build Nov. 22, 2011 (Huson et al., 2007; Huson et al., 2009)]. Each sequence was compared to the non-redundant (nr) database using BLASTX and results mapped to the NCBI Taxonomy Database. The second computational approach used VirusHunter software that identifies previously undescribed viruses via analysis of both nucleic acid and protein similarity (Felix et al., 2011; Loh et al., 2011; Zhao et al., 2011).
Pathogenic SIV infection is associated with an expanded enteric virome
We first analyzed the enteric virome of 44 rhesus monkeys housed at the NEPRC (22 monkeys infected intrarectally with pathogenic SIVmac251 and 22 SIV-uninfected monkeys, herein termed controls)(Figure 1A–B, S2A–B). SIV-infected and control rhesus monkeys were fed the same diet but housed separately. We analyzed fecal specimens 24 (Figure 1A, S2A) or 64 weeks (Figure 1B, S2B) after SIV infection; between these collection times 10 SIV-infected animals were euthanized for progressive AIDS. No control animals died.
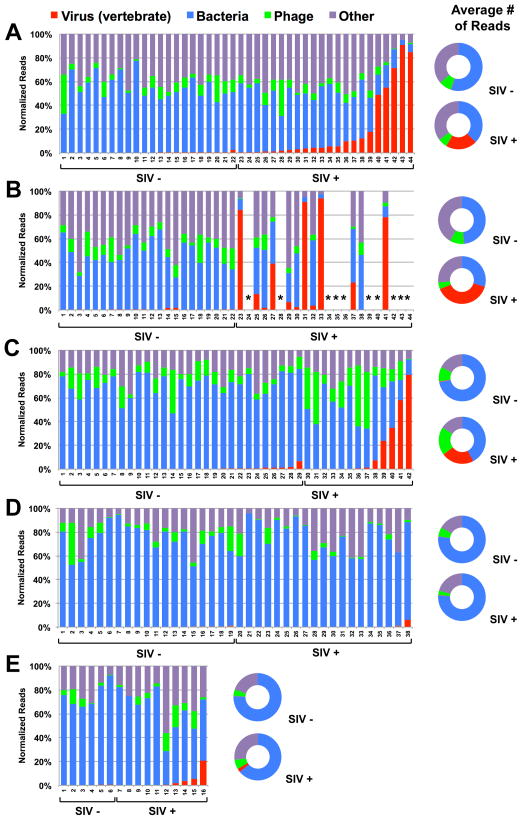
Figure 1. Taxonomic distribution of sequences identified in feces of SIV-infected (SIV+) and uninfected (SIV−) monkeys.
A–E graph the percentage of sequences obtained from fecal samples assigned by MEGAN to the indicated taxonomic groups. X-axis numbers refer to individual animals.
(A, B) Sequences from monkeys housed at NEPRC for 24 weeks (A) or 64 weeks (B) after intrarectal infection with SIVmac251. *euthanized for progressive AIDS 24 to 64 weeks after SIV infection.
(C) Sequences from monkeys housed at the TNPRC 23–64 weeks after intravaginal infection with SIVmac251.
(D) Sequences from SIV-infected and control vervet African green monkeys housed at NIH after intravenous infection with SIVagm90, SIVagmVer1 or after natural infection in the wild.
(E) Sequences from sabaeus African green monkeys housed at NEPRC and infected intravenously with SIVagmMJ8 or SIVagm9315BR.
(A–E) Flanking doughnut charts display the averaged values per kingdom for SIV+ or SIV- monkeys.
See also Figure S2
SIV infection was associated with a >10-fold increase in the number of sequences from viruses (p<0.0001) and a decrease in sequences from bacteria (p=0.003) at 24 weeks post-infection (Table 1, Figure 1A, S2A). There were no statistically significant SIV-associated changes in the total number of sequences from phages, alveolata (representing protists), viridiplantae (representing food sequences from plants), or other kingdoms and phyla (Figure 1A, S2A). Samples collected 40 weeks later revealed increases in viral sequences in most of the surviving animals that showed low numbers of viral sequences 24 weeks after SIV infection (e.g. compare animals 23, 31, and 33 between Figure 1A and 1B). Differences between SIV-infected and control monkeys, similar to those observed 24 weeks after SIV infection, were observed for both viral (p<0.0001) and bacterial (p=0.035) sequences 64 weeks after infection (Figure 1B, S2B). By 64 weeks post-infection, surviving SIV-infected monkeys also showed significant decreases in the number of phage (p=0.0320), alveolata (p=0.0183), and viridiplantae (p=0.0013) sequences compared to controls. These data suggest that pathogenic SIV infection is associated with significant expansion in the enteric virome.
To confirm these findings in pathogenically SIV-infected rhesus monkeys, we analyzed an independent cohort of 13 rhesus monkeys infected intravaginally with SIVmac251 and 29 controls housed at the TNPRC (Table 1, Figure 1C, S2C). SIV infection at the TNPRC was also associated with a significant increase in viral sequences (p=0.0420) and decrease in bacteria sequences (p=0.0019). In the TNPRC cohort, the SIV-infected monkeys showed significant increases in the number of phage (p=0.0133) sequences (Figure 1C, S2C). As for the 24 week time point in the NEPRC cohort, there were no significant changes in sequences from alveolata or viridiplantae or sequences from other kingdoms and phyla. The meaning of changes in phage sequences, which do not consistently track with pathogenic SIV infection, is uncertain. These results confirm that an expansion of the enteric virome is associated with pathogenic SIV infection in two independent cohorts of rhesus monkeys.
Non-pathogenic SIV infection is not associated with an expanded enteric virome
We next assessed whether the enteric virome changed during non-pathogenic SIV infection of African green monkeys (Table 1, Figure 1D–E, S2D–E). The vervet African green monkey cohort housed at the NIH was comprised of six monkeys infected intravenously with SIVagm90, two monkeys infected intravenously with SIVagmVer1, 11 monkeys naturally infected with SIV and 19 uninfected controls. The cohort of sabaeus African green monkeys housed at the NEPRC (Table 1, Figure 1E, S2E) was comprised of two monkeys infected intravenously with SIVagmMJ8, 8 monkeys infected intravenously with SIVagm9315BR and 6 uninfected control animals. Analysis revealed a decrease in phage sequences in the NIH cohort (p=0.0331) that was not observed in the NEPRC cohort, but no other significant SIV infection-associated changes (Figure 1D–E, S2D–E). Thus the expansion of the enteric virome observed during pathogenic SIV infection was not observed during non-pathogenic SIV infection. Importantly, these African green monkeys had been infected with SIV for a minimum of 3 years for the NIH cohort, and from 27 weeks (2 animals) to 2.6 years (8 animals) for the NEPRC cohort. Therefore, the lack of an increase in the enteric virome in these SIV-infected animals is not because they were infected for less time than pathogenically SIV-infected rhesus monkeys.
Viruses present in SIV-infected rhesus and African green monkeys
We next defined the nature of the enteric virome in SIV-infected and control monkeys using VirusHunter software (Figure 2A–E, Table S1). Using conservative criteria including the length of assembled contigs and the extents of divergence of sequences from the closest related virus in the NCBI non-redundant database, we detected at least 32 previously undescribed viruses in individual rhesus monkeys housed at the NEPRC alone (Figure 2A–B, Table S1). Certain viruses were found in multiple different animals, indicating shared exposure to enteric viruses. We did not count circoviruses in this estimate due to their ubiquity and diversity. Importantly, sequences from known insect (Dicistroviridae, Iflaviridae) or plant viruses, presumably derived from the diet, did not differ between SIV-infected and control animals (Figure 2A–E), indicating that our shotgun sequencing techniques do not artificially expand the enteric virome of SIV-infected rhesus monkeys.
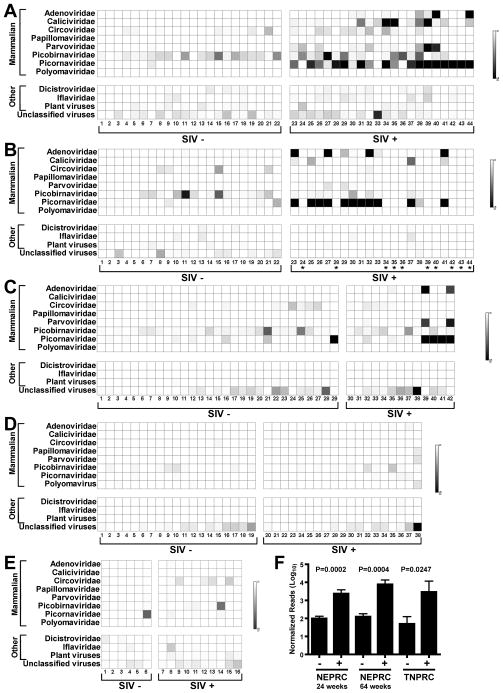
Figure 2. Distribution of virus sequences in rhesus and African green monkeys.
For these charts ‘mammalian’ indicates that sequences were most closely related to viruses that infect mammals. Viruses infecting non-mammals are referred to as ‘other’. ‘Unclassified virus’ includes all unclassified viruses, e.g. Chronic bee paralysis virus, Chimpanzee stool associated circular ssDNA virus, Circovirus-like genome RW-C, Circovirus-like genome CB-A, Rodent stool-associated circular genome virus, etc.
(A–E) The numbers below each chart and SIV infection status and panels are as in the Figure 1 legend.
(F) Comparison of the mean +/− SEM number of picornavirus sequences, after normalization for analysis using MEGAN, detected in the indicated cohorts of SIV-infected (+) and control (−) rhesus monkeys.
See also Table S1
Previously undescribed viruses identified here included five adenoviruses, three caliciviruses, one papillomavirus, eight members of the Parvoviridae (two parvoviruses, five dependoviruses, and one bocavirus), seven picobirnaviruses, seven members of the Picornavirales (three enteroviruses, three sapeloviruses, and one picornavirus), and one polyomavirus (Figure 2A–B, Table S1). Many SIV-infected rhesus monkeys at both NEPRC and TNPRC were shedding multiple potentially pathogenic viruses (Figure 2A–C, Table S1). The presence of multiple previously undescribed viruses, and of individual animals infected with multiple distinct viruses, was not regularly observed in control animals housed at the same location. In striking contrast, both cohorts of African green monkeys were relatively free of virus infection whether SIV-infected or not (Figure 2D, E).
As previously observed by others using classical virologic methods (Bailey and Mansfield, 2010; Oberste et al., 2002, 2007; Sasseville and Mansfield, 2010; Wang et al., 2007), picornaviruses were detected in both control and SIV-infected rhesus monkeys (Figure 2, Table S1). This allowed us to compare the number of sequences detected in pathogenic SIV-infected and control rhesus monkeys (Figure 2F). At both NEPRC and TNPRC there were more picornaviruses sequences in SIV-infected animals compared to controls (p=0.0002 and 0.0004, NEPRC animals at 24 or 64 weeks of infection, respectively; p=0.0247, TNPRC animals). No relationship was detected between picornavirus sequences and non-pathogenic SIV infection of African green monkeys (not shown). These data suggest a pathogenic SIV-associated failure to control picornavirus infection.
Identities of viruses in rhesus monkeys at the NEPRC
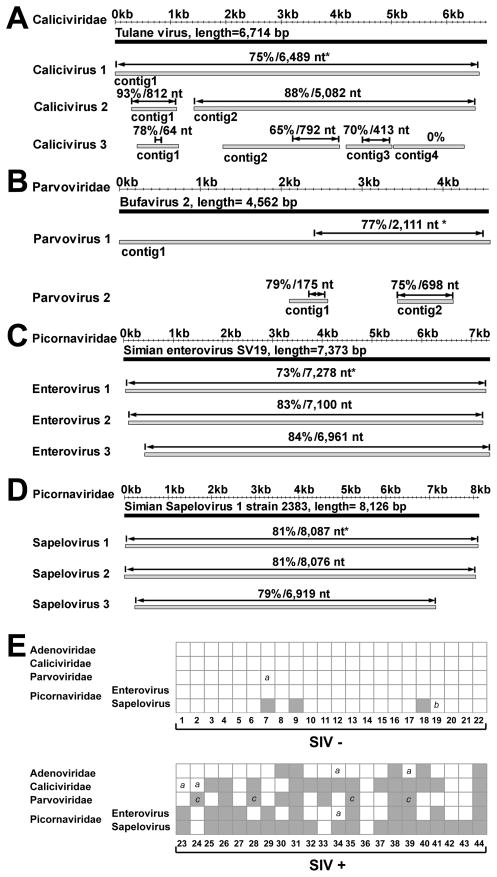
We next analyzed viruses present in the NEPRC cohort by assembling viral sequences from individual animals into contigs which we then compared to the most closely related virus present in the database (e.g. Figure 3A–D, 4A, Table S1, named using the convention ‘WUHARV-virus name-number’). Notably, some animals were shedding more than one virus of the same genus (Figures 2, 3, Table S1). We detected at least four adenoviruses (WUHARV Adenovirus 1–4, Figure 4A, not shown). We assembled portions of three calicivirus genomes (WUHARV Caliciviruses 1–3, Figure 3A, Table S3) with WUHARV Caliciviruses 1 and 2 most closely related to, but distinct from, the primate calicivirus Tulane (Farkas et al., 2008). For example WUHARV Calicivirus 1 shared only 75% nucleotide identity over a 6489 bp contig with Tulane and was phylogenetically distinct from Tulane (Figure S3). WUHARV Calicivirus 3 was quite distantly related to either Tulane virus or WUHARV Caliciviruses 1 and 2 (Figure 3A, not shown). We detected parvoviruses most closely related to Bufavirus 2, a recently described parvovirus (Phan et al.) (Figure 3B, S3). We assembled viral contigs covering most of the 7,000–8,000 bp genomes of several enteroviruses or sapeloviruses, both within the Picornaviridae (Figure 3C, 3D). WUHARV enteroviruses 1–3 share 73–84% nucleotide identity with simian enterovirus SV19, while WUHARV sapeloviruses 1–3 are 79–81% identical to simian sapelovirus 1 strain 2383 over essentially the entire genome (Figure 3C–D, S3). These data reveal a remarkable variety of viruses within the expanded enteric virome associated with pathogenic SIV infection.
Figure 3. Identification of enteric viruses in rhesus monkeys housed at the NEPRC.
For panels A–D grey bars represent assembled viral contigs, while black bars represent the genome of the most closely related known virus. *Indicates percent nucleotide identity over the designated length of the best aligned homologous region (indicated by double headed arrow) compared to the most closely virus genome. Animal numbers are as in Figure 1A.
(A) Contigs from WUHARV Caliciviruses 1 (animal 39), 2 (from an animal not included in the cohort), and 3 (animal 39) compared to Tulane calicivirus. Calicivirus 1 contig 1 derived from 879 sequences, length= 6,578 bp; Calicivirus 2 contig 1 derived from 16 sequences, length=812 bp; Calicivirus 2 contig 2 assembled from 120 sequences, length= 5,083 bp; Calicivirus 3 contig 1 assembled from 14 sequences, length= 750 bp; Calicivirus 3 contig 2 assembled from 67 sequences, length= 2,111 bp; Calicivirus 3 contig 3 assembled from 41 sequences, length= 832 bp; Calicivirus 3 contig 4 assembled from 38 sequences, length=1,273 bp.
(B) Contigs from WUHARV Parvovirus 1 (animal 39) and 2 (animal 35) compared with the sequence of Bufavirus 2. Parvovirus 1 contig 1 assembled from 375 sequences, length= 4,905 bp; Parvovirus 2 contig 1 representing 1 sequence, length= 470 bp; Parvovirus 2 contig 2 assembled from 6 sequences, length= 690 bp.
(C) Contigs from WUHARV Enterovirus 1 (animal 41), 2 (animal 39) and 3 (animal 33) compared with the sequence of Simian enterovirus SV19. Enterovirus 1 assembled from 1,084 sequences, length= 7,273 bp; Enterovirus 2 assembled from 758 sequences, length= 7,128 bp; Enterovirus 3 assembled from 406 sequences, length= 6,962 bp.
(D) Contigs from WUHARV Sapelovirus 1 (animal 42), 2 (animal 41) and 3 (animal 37) compared with the sequence of Simian Sapelovirus 1 strain 2383. Sapelovirus 1 assembled from 3,081 sequences, length= 8,059 bp; Sapelovirus 2 assembled from 2,711 sequences, length= 8,025 bp; Sapelovirus 3 assembled from 380 sequences, length= 6,872 bp.
(E) A chart showing the presence (grey box) of viral sequences in monkeys housed at NEPRC for 24 weeks as detected by PCR using virus-specific primers (Table S2). Numbers below the chart refer to the animals in Figure 1A. “a” = lack of detection of a virus likely due to the presence of a divergent virus; “b” = lack of detection of a virus for unknown reasons. “c” = detection of virus sequences in serum samples taken at the time of euthanasia for AIDs.
See also Figure S3 and Table S2
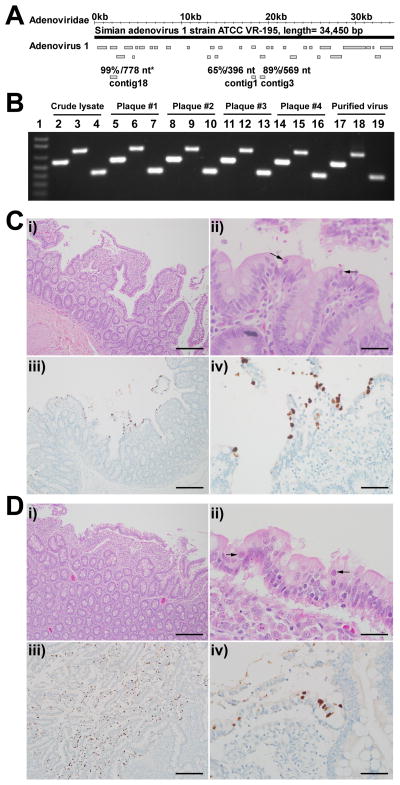
Figure 4. Enteric disease in SIV-infected rhesus monkeys at necropsy.
(A) The convention for showing viral contigs is as in Figure 3. Contigs from WUHARV Adenovirus 1 (animal #40) compared to the known virus Simian adenovirus 1 strain ATCC VR-195. These contigs were assembled from 1308 sequences.
(B) A gel showing PCR confirmation of WUHARV adenovirus 1 during amplification, plaque purification and cesium chloride gradient purification. The three PCR products for each sample (lanes 2–19) were derived from primers 4302c3f and 4302c3r, 4302c18f and 4302c18r, and 4302c1f and 4302c 1r respectively (Table S2). Lane 1, MW marker.
(C, D) Histopathology (top panels) and adenovirus immunohistochemistry (bottom panels) of the small intestine. Adenovirus infection was associated with villous atrophy and fusion (i) and sloughed epithelial cells that contained intranuclear adenoviral inclusions (ii, arrows). Adenovirus antigen could be localized to villous tip epithelium by immunohistochemistry (brown color of DAB chromagen, Mayer’s counterstain; iii and iv). Scale bars in i, iii = 0.5 mm. Scale bars in ii, iv=200 μm.
Detection of previously undescribed viruses by PCR and culture
We considered the possibility that NGS-detected viruses were actually present in all or most monkeys but the sequencing process was biased to detect viruses by pathogenic SIV infection. We therefore developed PCR assays to detect viruses for which we had large portions of the genome (Figure 3, Table S2), and used these independent assays to detect viruses (Figure 3E). In some cases contigs were so divergent that separate PCR assays were required to detect different viruses in the same group. For example, one PCR assay detected WUHARV Caliciviruses 1 and 2, while a different assay detected highly divergent WUHARV Calicivirus 3. Overall PCR analysis correlated well with NGS, agreeing in 62/69 cases (90%, Figure 3E), with some of the failures potentially related to the presence of viruses that were divergent from the viruses used to design PCR primers. PCR detected viruses in samples when as few as 1–2 viral sequences were detected by NGS. Compared to NGS, PCR detected 5/7 adenoviruses (failing to detect divergent adenoviruses in animals #34 and #39), 14/16 caliciviruses (failing to detect divergent caliciviruses in animals #23 and #24), 10/11 parvovirus genus members (failing to detect a divergent parvovirus in animal #7), 11/12 enterovirus genus members (failing to detect a divergent enterovirus in animal #34) and 22/23 sapelovirus genus members (failing to detect a non-divergent virus in animal #19 representing a true false negative). Importantly, PCR was negative for virus infection in a total of 151/151 cases for adenoviruses, caliciviruses, parvoviruses, enteroviruses, and sapeloviruses when NGS did not reveal a viral sequence. It remains possible that these viruses are present in additional animals not detected by PCR or NGS.
To further confirm NGS results, we cultured viruses from fecal samples. NGS data revealed (Figures 2A, 4A, Table S1) that multiple animals at NEPRC were potentially infected by previously undescribed adenoviruses. We therefore selected feces from animals #40 (60 adenovirus sequences), #44 (138 adenovirus sequences) and #30 (2 adenovirus sequences), as well as a fourth rhesus monkey not in this cohort (57 adenovirus sequences of 5758 unique reads) and sought to isolate viruses from them. We cultured five adenoviruses from these four animals (WUHARV Ad#1-5). These were sequentially plaque purified, amplified in culture, and isolated on cesium chloride gradients. We used PCR and sequencing to confirm that these viruses were those detected by NGS (WUHARV Ad1 shown in Figure 4A, and not shown). Together both PCR and culture analysis confirmed the presence of viruses detected by NGS in fecal samples from pathogenic SIV-infected animals.
Viruses in the expanded enteric virome are found in tissues and blood
To determine the clinical relevance of viruses detected by NGS, we evaluated the intestines of 12 necropsied SIV-infected animals (Figure 1B, Table 2). Five of 12 had intestinal pathology characteristic of cytomegalovirus, mycobacteria, or Balantidium (Table 2). Three animals (#23, 27, and 41) had high levels of adenovirus sequences prior to necropsy (Table 2), and these macaques, but not others in this necropsy cohort, exhibited adenovirus-associated enteritis by histologic examination (Figure 4C, D, panels i and ii). All had lesions in the jejunum and ileum (ileitis) while one also had lesions in the cecum (colitis). Immunohistochemistry confirmed the diagnosis of adenovirus ileitis (Figure 4C, D, panels iii and iv, Table 2), and colitis (not shown). Thus viruses detected in the fecal material of SIV-infected rhesus monkeys using NGS are associated with intestinal disease and epithelial damage in SIV-infected macaques.
Table 2.
Summary of Adenovirus detection and pathology in SIV-infected rhesus monkeys.
| Animal number | Number of Adenovirus reads1 | WUHARV Adenovirus1 | PCR screen1,2 | Adenovirus Enteritis3 | SI Adenovirus IHC3 | LI Adenovirus IHC3 | Other GI Pathologies3 |
|---|---|---|---|---|---|---|---|
| 23 | 889 | 1, others4 | Pos | Yes | Pos | Neg | Cytomegalovirus enteritis |
| 25 | 0 | n/a | Neg | No | Neg | Neg | No |
| 26 | 0 | n/a | Neg | No | Neg | Neg | No |
| 27 | 653 | 5, others4 | Pos | Yes | Pos | Neg | Balantidium sp. typhlitis |
| 29 | 14 | others4 | Neg | No | Neg | Neg | No |
| 30 | 1 | others4 | Neg | No | Neg | Neg | No |
| 31 | 0 | n/a | Neg | No | Neg | Neg | Mycobacterium avium enteritis_; Balantidium_ sp. colitis |
| 32 | 52 | others4 | Neg | No | Neg | Neg | No |
| 33 | 4 | others4 | Neg | No | Neg | Neg | No |
| 37 | 0 | n/a | Neg | No | Neg | Neg | No |
| 38 | 0 | n/a | Neg | No | Neg | Neg | Balantidium sp. colitis |
| 41 | 640 | others4 | Pos | Yes | Pos | Pos | Balantidium sp. typhlocolitis |
To further investigate the clinical relevance of viruses detected by NGS, we used virus specific PCR assays (Table S2) to determine whether viruses detected in the fecal material of SIV-infected rhesus monkeys (Figure 2A, 3E) were present in serum. We detected parvovirus (Figure 3E) in 4/10 serum samples taken at the time animals were euthanized for advanced AIDS between 24 and 64 weeks post infection. Sequence analysis of PCR amplicons demonstrated that parvoviruses present in fecal material (animals #24, #28, #35, #39) were also present in the serum of these four animals with advanced AIDS. This indicates that viruses detected in the fecal material of SIV-infected rhesus monkeys can invade tissues and enter the circulation, further supporting the concept that SIV-associated expansion of the enteric virus may contribute to disease.
Lack of an association between SIV infection and changes in the bacterial microbiome
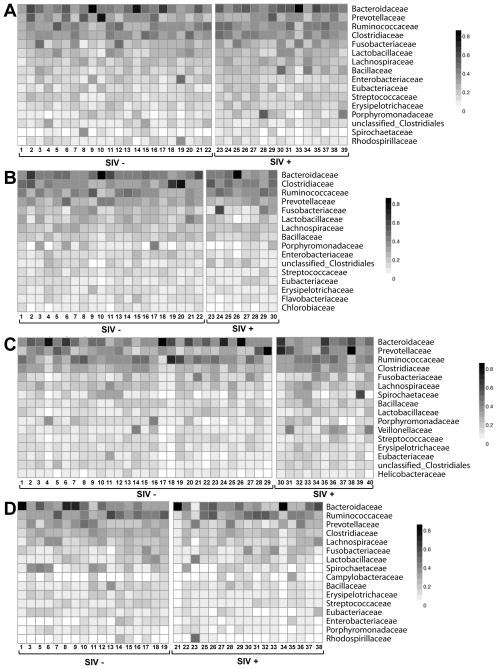
We next assessed the effects of SIV infection on the taxonomy of the bacterial microbiome (Fig. 5A–D). Our metagenomic data was comparable to published 16S rDNA-derived class-level data from SIV-infected and control macaques at TNPRC (McKenna et al., 2008), indicating that these distinct methods yield overall similar results (Figure S4). Rarefaction analysis revealed that all but a few samples with very high numbers of viral sequences were robust for analysis of bacterial diversity at the family level (Figure S4). Species accumulation curves indicated that all cohorts, except the NEPRC African green monkey cohort were robust for this analysis; further analysis excluded this cohort (Figure S4). We detected no consistent SIV-associated differences in bacterial family richness, evenness, or diversity (Legendre and Legendre, 1998). The significant (p=0.0345) SIV-associated difference in Shannon Diversity in the NEPRC cohort sampled 64 weeks post infection, was not observed in other pathogenic SIV-infected animals (NEPRC cohort 24 weeks post-infection, TNPRC cohort, (Figure S4). There was no difference in bacterial family evenness across cohorts (Figure S4). There were no significant differences between SIV-infected monkeys and uninfected controls in any cohort amongst the most-represented 20 bacterial families (Figure 5A–D). Additional analysis using principal component analysis as well as supervised and unsupervised random forest analysis (Yatsunenko et al.), showed no association between SIV infection and the bacterial microbiome. Further, we failed to find an association between SIV infection and either the genus- or species-level taxonomic structure of the bacterial microbiome. Thus, in contrast to our analysis of the virome, we detected no consistent SIV-infection associated differences in the family-level taxonomy of the bacterial microbiome.
Figure 5. Representative bacterial families in rhesus and African green monkey feces.
For A–D heat maps display the number of sequences assigned to specific bacterial families for individual animals in each cohort. The nature of SIV infection is as defined in the legend of Figure 1. (A, B) Sequences from pathogenic SIV-infected and control rhesus monkeys housed at the NEPRC (A) 24 weeks and (B) 64 weeks after SIV infection.
(C) Sequences from pathogenic SIV-infected and control rhesus monkeys housed at the TNPRC.
(D) Sequences from nonpathogenic SIV-infected and control vervet African green monkeys housed at the NIH.
See also Figure S4
DISCUSSION
Herein we report that pathogenic SIV infection is associated with a significant and unexpected expansion of the enteric virome detected using NGS of RNA and DNA. We documented a remarkable number of differences in the fecal virome between pathogenically SIV-infected monkeys, uninfected control monkeys, and monkeys infected with non-pathogenic SIV. These findings included increases in viral sequences, the presence of previously undescribed viruses, association of unsuspected adenovirus infection with intestinal disease and enteric epithelial pathology, and viremia with enteric parvoviruses in advanced AIDS. At least 32 previously undescribed viruses were detected from genera that cause diseases in mammalian hosts including adenoviruses, caliciviruses, parvoviruses, picornaviruses, and polyomaviruses. As our assignment of viral sequences to previously undescribed viruses was conservative, and as additional sequencing might detect additional viruses, we may have significantly underestimated both the size and the pathogenic potential of the enteric virome in SIV-infected animals. Furthermore, we may have missed viruses that infect the intestine but are shed at very low levels.
Application of standard diagnostic approaches such as PCR or culture would not have identified the breadth of divergent viruses detected here, and therefore would have underestimated both the potential causes of enteritis or systemic viral infection and the diversity of antigens which might contribute to enteropathy and immune activation. Our findings raise the interesting possibility that the nature of the enteric virome might be a prognostic indicator of HIV progression, and might contribute to AIDS pathogenenesis by damaging the intestinal epithelium to allow access of microbes, PAMPs, and viral antigens into tissues and the circulation to activate the immune system and stimulate lentivirus replication.
These data challenge the notion that abnormalities in the intestinal tract in pathogenic SIV-infected primates are due to direct effects of SIV or indirect effects of SIV on immune responses to enteric bacteria (Sandler and Douek). We suggest the distinct, but non-exclusive, hypothesis that immunocompromise during lentivirus infection is also associated with significant expansion of the enteric virome and that these viruses damage the intestine, as shown for adenoviruses in the present study. Such damage could provide access for bacterial PAMPs, or as shown here enteric viruses, into tissues and the circulation. It is already recognized that ‘bacterial’ and ‘viral’ contributions to intestinal pathology are not independent of each other. Clear synergies between the virome, bacteria, and host genes have been documented in murine systems (Bloom et al., 2011; Cadwell et al., 2010; Virgin and Todd, 2011). Importantly, it is not clear how bacterial PAMPs would explain the T cell activation characteristic of the systemic immune activation associated with AIDS progression. Our data suggest that T and B cell activation might be due to immune responses to unexpected viral antigens, as for example the parvovirus we detected in the circulation of a subset of animals. Unsuspected viral infections might also contribute to the high levels of IFN-alpha noted in the circulation of untreated AIDS patients. Searching for virus-specific T cell responses requires knowledge of the sequence of the viral proteins present, indicating the importance of sequencing the virome to define potential antigens that might drive immune activation in lentivirus-infected hosts.
A key observation is that many of the viruses we detected are RNA viruses and would not be detected in analyses of the microbiome utilizing DNA based sequencing of bacterial 16S rDNA or DNA-based shotgun sequencing. There has not been a complete analysis of the enteric microbiome at the RNA level to date, and to some extent the term ‘microbiome’ has been used to refer to bacteria alone rather than all taxa of life present in the intestinal wall or intestinal contents. In addition to viruses, for example, commensal fungi and bacteria have been associated with colitis (Bloom et al., 2011; Iliev et al., 2012). Indeed a broad range of organisms can interact with host genes to alter the phenome of the host (Virgin and Todd, 2011; Virgin et al., 2009). For example ‘virus plus gene’ interactions can induce human-like pathology in mice, indicating that complex interactions between the enteric virome and host genes may contribute to a range of phenotypes (Cadwell et al., 2008; Cadwell et al., 2010; Virgin and Todd, 2011; Virgin et al., 2009). The need for a broad and unbiased assessment of the DNA and RNA-defined microbiome in association with enteric disease is clearly indicated by our detection of expansion of the virome associated with pathogenic SIV infection.
The complexity of the enteric virome
An important issue raised by our findings is how to taxonomically assign viral sequences when only portions of the viral genome are present. When complete viral genomes are available, their assignment to family, genus, species and strain can be made based on historical criteria in initial publications and then codified by the International Committee on the Taxonomy of Viruses (ICTV at http://ictvonline.org/). As we did not have complete genomes for the 32 viruses we identified here, we elected to report in Figures 3 and S3 and Table S1 (and to target by PCR) a group of viruses for which we had significant portions of the genome and could use very conservative criteria for relatedness between viruses. As sequence depth increases and assembly becomes more robust, the availability of more viral genomes and more complete viral genomes will allow clearer assignment of viral genomes and assessment of the breadth of the virome. An important conceptual issue herein is that viral pathogenesis and virulence is often conferred by single or a few nucleotide changes. Furthermore, many of the immunocompromised monkeys studied here were shedding multiple potentially pathogenic viruses. Thus it will be a major task to select the viral agents to be studied to understand the contribution of the complex enteric virome to disease pathogenesis.
SIV and the enteric bacterial microbiome
We failed to find a clear association between pathogenic SIV-infection and multiple independent measures of family-level bacterial diversity and population structure. We also examined this question at the genus, and species level but note that these data sets were less robust as judged by rarefaction analysis of individual animals. These data present a striking contrast with expansion of the enteric virome we document in monkeys infected with pathogenic SIV. These analyses are consistent with the single other study of macaques and SIV indicating that there are no major family-level alterations in fecal bacteria associated with pathogenic SIV infection (McKenna et al., 2008). This conclusion comes with significant caveats. Firstly, some samples in our study with very high numbers of viral sequences failed rarefaction, leaving open the possibility that when virus infection is very high there are changes in enteric bacteria. Further, the number of sequences analyzed here allowed assessment of family-level taxonomy but not a detailed assessment at the genus, species or strain level. In addition, fecal material may not reflect the populations of bacteria that adhere to the intestinal mucosal (Nava et al., 2011). Further analyses of possible SIV-associated changes in the bacterial microbiome, and of relationships between the virome and the microbiome, will therefore require generation of sequence libraries large enough to support analysis of the bacterial microbiome at the genus, species, and strain level.
Implications for AIDS Pathogenesis
Discovery of the expansion of the enteric virome in nonhuman primates infected with pathogenic SIV, but not with non-pathogenic SIV, has profound implications for understanding AIDS pathogenesis in these animals and indicates the need for similar studies in human AIDS. Our data are consistent with a model in which immunosuppression results in increased levels of enteric viral infection which, in a feed-forward manner, contributes to AIDS via damage to the intestinal mucosa and induction of systemic immune activation that accelerates AIDS progression. The pathogenetic potential of the enteric virome, exemplified by animals with enteritis associated with adenovirus infection or parvovirus viremia, is not fully understood based on this initial study. By sequencing both RNA and DNA and by using metagenomic approaches, rather than focusing on bacterial 16S rDNA analysis, we have documented a previously undescribed set of viruses associated with clinical AIDS progression in rhesus monkeys. Since these viruses include many potential pathogens, studies of HIV and SIV pathogenesis should take them into account as possible contributors to disease progression. This provides substantial opportunity to explain and eventually intervene in the processes that lead to AIDS clinical disease progression. Furthermore, our data suggest that expansion of the enteric virome may be useful as a marker for rapidly progressive disease. Future studies will directly investigate the role of both RNA and DNA components of the metagenome in AIDS pathogenesis in both nonhuman primates and humans. Such studies will lead to a more detailed understanding of AIDS enteropathy and the molecular basis of systemic immune activation that is associated with progression to AIDS.
EXPERIMENTAL PROCEDURES
Nucleic acid preparation and shotgun sequencing
Frozen stool was resuspended in PBS, centrifuged, and the supernatant passed through a 0.45 μm filter. Total RNA and DNA was isolated from the filtrate, reverse transcribed and PCR amplified using bar-coded primers (Wang et al., 2003). Amplification products were sequenced on the 454 GS FLX Titanium platform (454 Life Sciences). See supplemental methods for details.
Detection and analysis of viral sequences using custom bioinformatic pipeline
Sequences were analyzed using VirusHunter software as described (Felix et al., 2011; Loh et al., 2011; Loh et al., 2009; Presti et al., 2009; Zhao et al., 2011). Briefly, sequences were assigned to individual samples using barcode sequences, primer sequences were trimmed, and sequences were clustered using CD-HIT (Li and Godzik, 2006) to remove redundant sequences (95% identity over 95% sequence length). The longest sequence from each such cluster was chosen as the representative unique sequence, and entered into the analysis pipeline. Sequences were masked by RepeatMasker (http://www.repeatmasker.org); those lacking at least 50 consecutive non-”N” nucleotides or having >40% of their length masked were removed (filtered). Filtered high quality unique non-repetitive sequences were sequentially compared against (i) the human genome using BLASTn; (ii) GenBank nt database using BLASTn; and (iii) GenBank nr database using BLASTX (Altschul et al., 1990). Minimal e-value cutoffs of 1e−10 and 1e−5 were applied for BLASTn and BLASTX respectively (Bench et al.; Wommack et al.). Sequences were phylotyped as human, mouse, fungal, bacterial, phage, viral, or other based on the top BLAST hit. Sequences without any significant hit in any database were designated as unassigned. Sequences aligning to both a virus and another kingdom (e.g. bacteria or fungi) with the same e value were classified as ambiguous. All eukaryotic viral sequences were further classified into viral families based on the taxonomy ID of the best hit.
Assembly of viral contigs and virus comparison analysis
All viral sequences and unassigned sequences (and 5 longest sequences similar to these sequences) from each sample were assembled into contigs using Newbler (454 Life Sciences) with default parameters. If a sample was sequenced multiple times, all available sequence data was used to optimize contig assembly. The longest assembled contig belonging to a given genus was analyzed first as the ‘representative’ contig. To compare viruses across multiple animals, contigs (and sequences if no contigs were obtained from a sample) were compared with this ‘representative’ contig. Sequences sharing 98% nucleotide identify or higher over the aligned region with the ‘representative’ contig were considered to be the same virus, and removed from further analysis. This process was sequentially repeated for all remaining contigs until all sequences were classified. If two contigs or sequences from a single sample were homologous to different regions of a known viral genome, we made the conservative assumption that that only a single virus was present. Unique viral contigs selected in this manner were queried against the NCBI nt database and the most closely related complete viral genome was selected as the reference genome. For adenoviruses, different NGS sequences and contigs shared the highest homology with different known viruses. Two out of the 3 contig sequences used for designing primers shared highest homology to Simian adenovirus 1 strain ATCC VR-195, which was therefore selected as reference genome. If no nucleotide level homology was detected, viral contigs were queried for protein homology against the NCBI nr database and the most related viral genome was identified.
Metagenomic analysis using MEGAN
Individual sequences were analyzed using BLASTX (version 2.2.22+) on a customized server with ~1700 available processor slots and a memory range of 2–32 GB per node. Sequences were compared by BLASTX to the NCBI nr database version 06/06/2011. Results with an e-value e−10 were stored and used for taxonomic assignment using the Lowest-Common Ancestor (LCA) algorithm in MEGAN v. 4.62.3 build 22 Nov 2011. The following LCA parameters were used for taxonomic assignment: Min Support: 5, Min Score: 35, Top percent 10, Win Score: 0, Min Complexity 0. This generated sample-specific RMA files containing all of the taxonomic assignment information for each sample to be used for downstream analysis. Global metagenome comparisons using all sequences assigned to all taxa were completed for each cohort. These comparisons used MEGAN’s normalization protocol enabling inter-sample comparison. Additionally, sequences in specific taxa (bacteria, viruses, or phage) were isolated and processed through MEGAN using the same parameters to independently analyze these taxa without effects of global normalization. Summarized sequence counts per taxa were exported for subsequent statistical analysis using GraphPad Prism version 5.0d.
PCR detection of viruses
Primers (Table S2) were designed to amplify regions conserved between WUHARV adenoviruses 1–5, caliciviruses 1–2, calicivirus 3, parvoviruses 1–2, enteroviruses 1–3, sapeloviruses 1–3 and related viral genomes. Primer sensitivity was evaluated using libraries with high or low numbers of adenovirus, calicivirus, parvovirus, enterovirus or sapelovirus sequences, while primer specificity was evaluated using libraries with high numbers of unrelated virus sequences, as well as virus sequences from related genera. Libraries generated from stool samples were diluted 10 fold and screened (n=2) by PCR for presence of viruses. There was concordance in all duplicate tests. See supplemental methods for details.
Isolation and detection of WUHARV adenoviruses
Filtrates of stool samples were used to inoculate adenovirus-permissive cells, viruses were plaque purified twice on Per55K cells, and then used to generate virus stocks and CsCl-purified virus. To detect adenoviruses, primers (Table S2) were designed to amplify regions from WUHARV adenoviruses (1–5) from contigs with a range of relatedness to the reference genomes and then visualized using EtBr on a 0.8% agarose gel. See supplemental methods for details.
Assays and necropsy of SIV-infected rhesus monkeys
Serum levels of LPS binding protein (LBP) were quantitated by ELISA (Antibodies Online). Twelve animals housed at the NEPRC (Figure 1B) were subjected to complete necropsy within two hours of death and representative histologic sections of all major organs were analyzed for pathology. Immunohistochemistry using an adenovirus specific antibody was used to detect infected cells. See supplemental methods for details.
GenBank accession numbers
Sequence data from each animal were uploaded to the MG-RAST server (version 3.12). The sequences of virus contigs presented in Figure 3 and Figure S2 have GenBank accession numbers as follows: WUHARV Calicivirus 1 (JX627575), WUHARV Parvovirus 1 (JX627576), WUHARV Enterovirus 1 (JX627570), WUHARV Enterovirus 2 (JX627571), WUHARV Enterovirus 3 (JX627572), WUHARV Sapelovirus 1 (JX627573), WUHARV Sapelovirus 2 (JX627574).
Statistical analysis
For analysis of sequence numbers after normalization the data were log10 transformed prior to statistical analysis. P-values were derived using the nonparametric Mann-Whitney test. P-values < 0.05 are considered significant. For analysis of bacterial families in Figure 5, we utilized one-way ANOVA with a Bonferroni correction to correct for multiple comparisons.
Supplementary Material
01
02
Highlights.
- The enteric virome expands during pathogenic, but not non-pathogenic, SIV infection.
- Pathogenic SIV-infected rhesus monkeys shed multiple enteric viruses.
- Viruses are associated with intestinal pathology during SIV infection.
- Metagenomic analysis of the enteric virome is important to study AIDS pathogenesis.
Acknowledgments
This work was supported by: Project 10 of U54 AI057160-08 to DW for development of VirusHunter software, was initially funded by the National Center for Research Resources 1R01 RR032309 and is currently supported by the Office of Research Infrastructure Programs OD11170-02 to HWV and DB, Crohn’s and Colitis Foundation Grant 3132 to HWV and TS, grants AI066305, AI066924, AI078526, and AI095985 to DB, AI65335 to JES, and grant 8P51OD011103-5 to ADM and JK. We would like to thank Angela Carville and Vanessa Hirsch for assistance with these experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. JMolBiol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bailey C, Mansfield K. Emerging and reemerging infectious diseases of nonhuman primates in the laboratory setting. VetPathol. 2010;47:462–481. doi: 10.1177/0300985810363719. [DOI] [PubMed] [Google Scholar]
- Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HW. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- Bench SR, Hanson TE, Williamson KE, Ghosh D, Radosovich M, Wang K, Wommack KE. Metagenomic characterization of Chesapeake Bay virioplankton. Applied and environmental microbiology. 2007;73:7629–7641. doi: 10.1128/AEM.00938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol. 2012;30:149–173. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Price DA, Douek DC. HIV disease: fallout from a mucosal catastrophe? Nat Immunol. 2006a;7:235–239. doi: 10.1038/ni1316. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. NatMed. 2006b;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Silvestri G, Douek DC. Nonprogressive and progressive primate immunodeficiency lentivirus infections. Immunity. 2010;32:737–742. doi: 10.1016/j.immuni.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, Barclay GR, Smedley J, Pung R, Oliveira KM, et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 2010;6:e1001052. doi: 10.1371/journal.ppat.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T, Sestak K, Wei C, Jiang X. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J Virol. 2008;82:5408–5416. doi: 10.1128/JVI.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Belicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. GenomeRes. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Mitra S, Auch AF, Schuster SC. Methods for comparative metagenomics. BMC Bioinformatics. 2009;10(Suppl 1):S12. doi: 10.1186/1471-2105-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Legendre L. Numerical Ecology. Amsterdam: Elsevier Science B.V; 1998. Vol Second English Edition. [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Loh J, Zhao G, Nelson CA, Coder P, Droit L, Handley SA, Johnson LS, Vachharajani P, Guzman H, Tesh RB, et al. Identification and sequencing of a novel rodent gammaherpesvirus that establishes acute and latent infection in laboratory mice. J Virol. 2011;85:2642–2656. doi: 10.1128/JVI.01661-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J, Zhao G, Presti RM, Holtz LR, Finkbeiner SR, Droit L, Villasana Z, Todd C, Pipas JM, Calgua B, et al. Detection of novel sequences related to african Swine Fever virus in human serum and sewage. J Virol. 2009;83:13019–13025. doi: 10.1128/JVI.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, Lozupone CA, Hamady M, Knight R, Bushman FD. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISMEJ. 2011;5:627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste MS, Maher K, Pallansch MA. Molecular phylogeny and proposed classification of the simian picornaviruses. J Virol. 2002;76:1244–1251. doi: 10.1128/JVI.76.3.1244-1251.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste MS, Maher K, Pallansch MA. Complete genome sequences for nine simian enteroviruses. J GenVirol. 2007;88:3360–3372. doi: 10.1099/vir.0.83124-0. [DOI] [PubMed] [Google Scholar]
- Pandrea I, Gaufin T, Brenchley JM, Gautam R, Monjure C, Gautam A, Coleman C, Lackner AA, Ribeiro RM, Douek DC, et al. Cutting edge: Experimentally induced immune activation in natural hosts of simian immunodeficiency virus induces significant increases in viral replication and CD4+ T cell depletion. J Immunol. 2008;181:6687–6691. doi: 10.4049/jimmunol.181.10.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Vo NP, Bonkoungou IJ, Kapoor A, Barro N, O’Ryan M, Kapusinszky B, Wang C, Delwart E. Acute diarrhea in West-African children: diverse enteric viruses and a novel parvovirus genus. J Virol. 2012 doi: 10.1128/JVI.01427-12. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presti RM, Zhao G, Beatty WL, Mihindukulasuriya KA, Travassos da Rosa AP, Popov VL, Tesh RB, Virgin HW, Wang D. Quaranfil, Johnston Atoll and Lake Chad viruses are novel members of the family Orthomyxoviridae. J Virol. 2009;83:11599–11606. doi: 10.1128/JVI.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nature reviews Microbiology. 2012;10:655–666. doi: 10.1038/nrmicro2848. [DOI] [PubMed] [Google Scholar]
- Sasseville VG, Mansfield KG. Overview of known non-human primate pathogens with potential to affect colonies used for toxicity testing. J Immunotoxicol. 2010;7:79–92. doi: 10.3109/15476910903213521. [DOI] [PubMed] [Google Scholar]
- Sodora DL, Allan JS, Apetrei C, Brenchley JM, Douek DC, Else JG, Estes JD, Hahn BH, Hirsch VM, Kaur A, et al. Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. NatMed. 2009;15:861–865. doi: 10.1038/nm.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell. 2011;147:44–56. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- Wang D, Urisman A, Liu YT, Springer M, Ksiazek TG, Erdman DD, Mardis ER, Hickenbotham M, Magrini V, Eldred J, et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tu X, Humphrey C, McClure H, Jiang X, Qin C, Glass RI, Jiang B. Detection of viral agents in fecal specimens of monkeys with diarrhea. J MedPrimatol. 2007;36:101–107. doi: 10.1111/j.1600-0684.2006.00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Bhavsar J, Ravel J. Metagenomics: read length matters. Applied and environmental microbiology. 2008;74:1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Droit L, Tesh RB, Popov VL, Little NS, Upton C, Virgin HW, Wang D. The genome of Yoka poxvirus. J Virol. 2011;85:10230–10238. doi: 10.1128/JVI.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02