Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2 (original) (raw)
. Author manuscript; available in PMC: 2013 Aug 14.
Summary
The microtubules that comprise mitotic spindles in animal cells are nucleated at centrosomes and by spindle assembly factors that are activated in the vicinity of chromatin. Indirect evidence also has suggested that microtubules might be nucleated from pre-existing microtubules throughout the spindle, but this process has not been observed directly. Here, we demonstrate microtubule nucleation from the sides of existing microtubules in meiotic Xenopus egg extracts. Daughter microtubules grow at a low branch angle and with the same polarity as mother filaments. Branching microtubule nucleation requires gamma-tubulin and augmin and is stimulated by GTP-bound Ran and its effector TPX2, factors previously implicated in chromatin-stimulated nucleation. Because of the rapid amplification of microtubule numbers and the preservation of microtubule polarity, microtubule-dependent microtubule nucleation is well suited for spindle assembly and maintenance.
Introduction
Mitotic and meiotic spindles are built from dynamic microtubules arranged in two half-spindles with anti-parallel overlap in the center. Generating and maintaining this array requires controlling microtubule nucleation both in space and time. The first described and best understood mechanism involves microtubule nucleation from centrosomes (Brinkley, 1985), which is thought to be mediated by γ-tubulin ring complexes (γ-TuRC) tethered to pericentriolar proteins (Keating and Borisy, 2000; Kollman et al., 2010; Moritz et al., 2000; Wiese and Zheng, 2000; Wiese and Zheng, 2006). However, plant cell mitosis and animal egg meiosis occur without centrosomes (Dumont and Desai, 2012; Zhang and Dawe, 2011), and animal cell mitosis can occur normally after the centrosomes have been ablated (Khodjakov et al., 2000; Megraw et al., 2001). Thus, nucleation of spindle microtubules does not always rely on centrosomes.
A pathway for generating acentrosomal microtubules emerged from the finding that nuclei lacking centrosomes injected into Xenopus eggs promoted microtubule nucleation (Karsenti et al., 1984). This pathway was later recapitulated in egg extracts using chromatin-coated beads that stimulated spindle formation in Xenopus egg extracts (Heald et al., 1996; Karsenti and Vernos, 2001). Subsequent work revealed that this chromatin-stimulation of microtubule nucleation is mediated by the small GTPase Ran (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999), which, in its GTP state, releases spindle assembly factors from importins (Gruss et al., 2001; Wiese et al., 2001). The Ran nucleotide exchange factor (RCC1) localizes to chromatin, resulting in a Ran-GTP gradient (Caudron et al., 2005; Kalab et al., 2006; Kalab et al., 2002) which is thought to promote microtubule nucleation in the vicinity of chromatin (Athale et al., 2008; Caudron et al., 2005; Karsenti and Vernos, 2001). Microtubules do not emerge directly from chromatin in this pathway; rather they form near chromatin, typically as large bundles whose precise origin is unclear. The primary downstream microtubule nucleator in the Ran pathway is thought to be γ-TuRC (a large protein complex consisting of γ-tubulin (γ-TB) and several associated subunits), although other factors may also contribute (Groen et al., 2009). The details of how g-TuRC is linked to the Ran-GTP pathway is not completely clear although one of the intermediate effectors is the coiled coil protein TPX2, which is released from importins by Ran-GTP and plays an important role in microtubule generation in both acentrosomal (Gruss et al., 2001) and centrosomal spindles (Kufer et al., 2002). In C. elegans, TPX2 levels determine the size of the first mitotic spindle, suggesting it participates in a rate-limiting step for spindle assembly (Greenan et al., 2010). However, the mechanism by which TPX2 leads to chromosomal-stimulated microtubule nucleation remains unclear.
Recent work also has shown that new microtubules are generated within the body of metazoan somatic spindles (Brugues et al., 2012; Mahoney et al., 2006). Augmin, a protein complex discovered based upon its ability to target γ-TB to spindle microtubules (Goshima et al., 2007), has been implicated in this process. Augmin depletion lowers microtubule density within the spindle and the anaphase midzone, destabilizes kinetochore fibers, and results in chromosome misalignment (Goshima et al., 2008; Goshima et al., 2007; Hotta et al., 2012; Lawo et al., 2009; Nakaoka et al., 2012; Petry et al., 2011; Uehara and Goshima, 2010; Uehara et al., 2009; Zhu et al., 2008). Augmin also plays a role in acentrosomal spindle formation by contributing to the microtubule mass in the spindle, especially during the initial phase of assembly, and by stabilizing the bipolar spindle shape (Petry et al., 2011). Augmin was proposed to recruit γ-TuRC to the side of a pre-existing microtubule and thus initiate the nucleation of a new microtubule (Goshima et al., 2008), analogous to how Arp2/3 complex nucleates actin filaments by binding to a pre-existing actin filament (Blanchoin et al., 2000). However, this process, which we term branching microtubule nucleation, has not been observed directly in a spindle or in a metazoan organism. Microtubule nucleation from the sides of pre-existing microtubules has only been observed in fission yeast (antiparallel growth; (Janson et al., 2005)) and plant cells (parallel growth at a ~ 42 degree angle; (Murata et al., 2005)), but the factors that are responsible for docking γ–TB to the side of pre-existing MTs in these systems have not been determined and these processes have not been recapitulated using in vitro systems.
A possible reason why branching MT nucleation has not been visualized during mitosis/meiosis is because the very high microtubule density in the spindle might preclude direct observation of such events. Here, we have succeeded in visualizing branching MT nucleation using meiotic Xenopus egg extracts and total internal reflection (TIRF) microscopy. This in vitro assay has allowed us to analyze the angles of branching microtubule nucleation, the contribution of branching nucleation to overall microtubule formation in the extract, and the molecular factors that underlie this process.
Results
Visualization of branching microtubule nucleation in meiotic Xenopus egg extracts
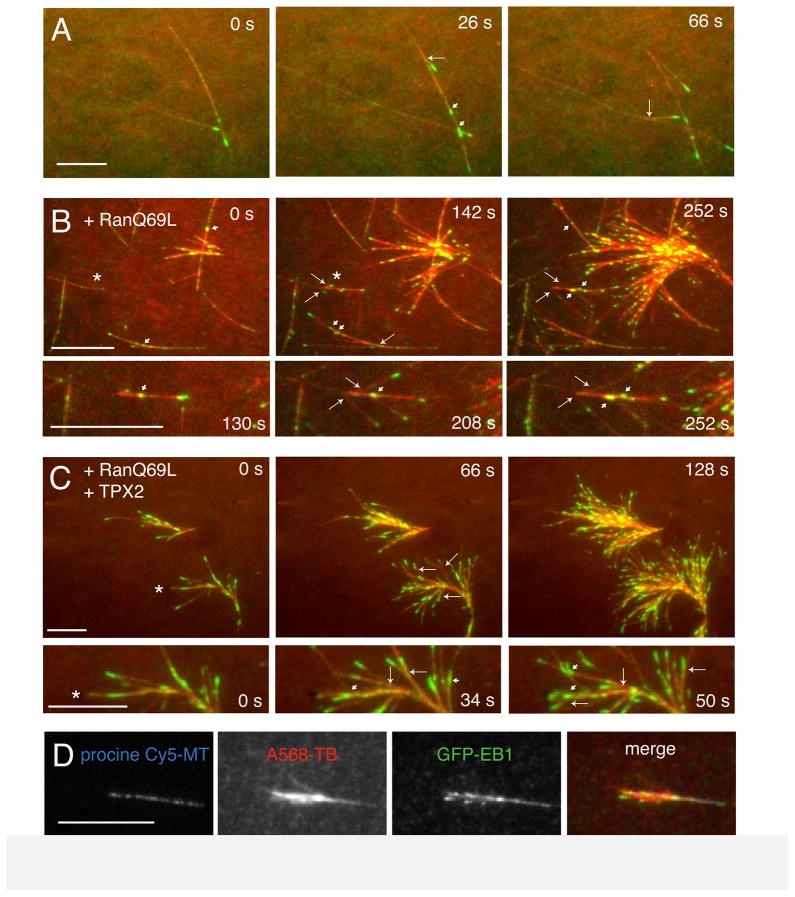
To visualize microtubule nucleation as it may occur in a metazoan spindle, we used cell-free extracts from Xenopus oocytes arrested at meiosis II, which accurately recapitulate assembly of the acentrosomal egg meiosis II spindle (Heald et al., 1996; Sawin and Mitchison, 1991). GFP-EB1, which tracks the growing microtubule plus ends and thus highlights new microtubule growth, and Alexa568-labeled tubulin were added to the extract and microtubule formation/growth was visualized near the glass surface by total internal reflection (TIRF) microscopy. Observation of microtubule dynamics, however, was hampered by their rapid transport across the glass surface by cytoplasmic dynein, the dominant motor activity in the extract. We therefore inhibited microtubule gliding with vanadate. Microtubules typically displayed stable (neither growing or shrinking) microtubule minus ends and growing microtubule plus ends marked by GFP-EB1 at the tip; the velocity of plus end growth was 13.4 ± 1.3 μm/min (mean and sd, n = 30), which is similar to previous studies using Xenopus extracts (Tirnauer et al., 2004). Catastrophies (transition to a rapidly shrinking phase) were rarely observed. Occasionally, a new microtubule was observed (through the formation of a new GFP-EB1 spot) to form at the side of an existing microtubule and then to grow either along or at a shallow angle to the template microtubule (Fig. 1A; Movie S1). This assay provides the first unambiguous observation of branching MT nucleation in a metazoan system and allowed further analysis of the factors involved.
Figure 1. Branching microtubule nucleation is stimulated by Ran and TPX2 in meiotic Xenopus egg extracts.
(A) Branching microtubule nucleation in Xenopus egg meiotic extracts without added Ran or TPX2. EB1-GFP (green) was added to the extract to follow the microtubule plus ends and thus identify locations of new microtubule growth. Alexa568-labelled bovine brain tubulin (red) was added to visualize microtubules. Sodium orthovanadate was added to prevent dynein-mediated sliding of microtubules along the glass and allow better observation of branching microtubules. Large arrows indicate nucleated microtubules that emerge a clear angled branch from the mother microtubule. Short arrows indicate nucleated microtubules that grow along the length of the mother microtubule. The time of this sequence is shown in seconds. Scale bar, 5 μm. See Movie S1. (B) Branching nucleation is stimulated in the presence of a constitutively active Ran mutant (Ran Q69L). The asterisk indicate a region that is enlarged in the bottom panels. Scale bar, 5 μm. See Movie S2. (C) Microtubule nucleation in an extract containing both RanQ69L and TPX2, leading to branched-fan like structures in the absence of microtubule gliding. The asterisk indicates a region that is enlarged in the bottom panels. Scale bar, 5 μm. See Movie S3. (D) Branching occurs from an exogenously added template microtubule. GMPCPP-stabilized, Cy5-labeled pig brain microtubules were attached to a passivated glass coverslip (to prevent binding of endogenous microtubules), and a reaction mixture of Xenopus extract, RanQ69L, TPX2, EB1-GFP and alexa568-labelled bovine brain tubulin was added. New microtubule growth (EB1-GFP spots) can be seen emerging from the template microtubule. Scale bar, 10 μm. See also Figure S1.
Branching microtubule nucleation is promoted by RanGTP and TPX2
In order to assay branching microtubule nucleation in a more robust manner, we sought factors that might stimulate this process. RanGTP, trapped in this nucleotide state by the non-hydrolyzing mutant RanQ69L, induces formation of microtubule asters and mini-spindles in the absence of centrosomes or chromatin (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999) and microtubule formation occurs in an apparent autocatalytic manner after RanGTP addition (Clausen and Ribbeck, 2007). Our previous work showing that augmin immunodepletion delayed the formation of RanGTP asters and mini-spindles also suggested a possible connection between Ran and augmin-mediated microtubule nucleation (Petry et al., 2011). To test this idea further, we added recombinant RanQ69L to the extract system. Initially, a few isolated microtubules appeared, followed by nucleation of new microtubules along the length of the initial template microtubules (Fig. 1B; Movie S2). By this branching mechanism RanGTP dramatically enhanced branching microtubule nucleation in the Xenopus egg extract. An important Ran effector for spindle assembly is TPX2 (Gruss et al., 2001). The combined addition of recombinant RanQ69L and TPX2 further stimulated branching microtubule nucleation (Fig. 1C, Movie S3). In the absence of microtubule gliding, the junctions between daughter and mother microtubules remained intact for >10 min. This resulted in the consistent appearance of large fan-shaped microtubule structures composed of a tight bundle of microtubules at the base and an expanding network of branching microtubules all pointing in a similar direction. Addition of TPX2 alone promoted microtubule branching (Fig. S1D), although qualitatively there were more single microtubules and the fan-shaped microtubule structures that formed were less dense than when TPX2 and RanQ69L were combined together.
The assay conditions described above utilized vanadate, which prevented dynein from pulling newly nucleated daughter microtubules away from the mother microtubule. However, branching nucleation events in the presence of RanQ69L or RanQ69L/TPX2 were clearly observed in the absence of vanadate (Fig. S1A and B; Movies S4 and S5), indicating that this inhibitor was not essential for the nucleation reaction. However, the “fans” were less organized in the absence of vanadate due to their disruption by microtubule gliding. We also could observe branching and fan-shaped microtubule structures by blocking dynein gliding with CC1, an inhibitory fragment of the p150 subunit of dynactin (Quintyne et al., 1999) (Fig. S1C).
Branching microtubule nucleation from an exogenous template microtubule
We next tested whether branching microtubule nucleation can occur on an exogenously added, stabilized microtubule. For this purpose, GMPCPP-stabilized, biotinylated porcine brain microtubules were tethered via an anti-biotin antibody to the surface of a passivated glass coverslip. Upon addition of meiotic Xenopus egg extract complemented with RanQ69L and TPX2, robust branching nucleation occurred exclusively along the length of the stabilized microtubules, but not on the passivated glass surface (Fig. 1D). This further demonstrates that RanGTP and TPX2 can induce the formation of daughter microtubules from a single parent microtubule, resulting in a tight bundle of microtubules with the same polarity. In addition, this result indicated that the parent microtubule need not be dynamic and indeed can be from another species.
The kinetics and properties of branching microtubule nucleation
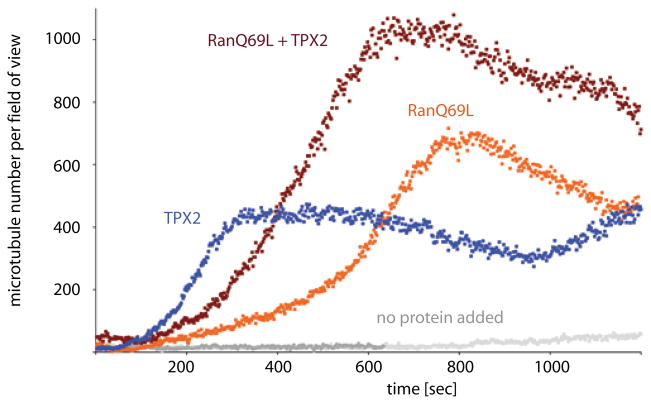
The qualitative observations from the microscopy experiments described above demonstrate that addition of exogenous Ran-GTP and TPX2 stimulate branching microtubule nucleation. We next sought to determine how these factors influence the kinetics of microtubule nucleation. An individual microtubule is marked by a single EB1-GFP spot at its growing plus tip, and thus counting the number of EB1-GFP spots using an automated algorithm provides a direct measure of microtubule number over time. Application of this method to TIRF imaging only counts microtubules near the coverslip, but non-quantitative confocal and widefield imaging performed on similar reactions (not shown) suggested this reflects the total population. In the wildtype extract, microtubule numbers increased very slowly over time (Fig. 2). Addition of RanQ69L dramatically stimulated the appearance of microtubules compared to the control extract; the kinetics revealed an initial lag phase followed by a sigmoidal increase in microtubule number. This result is in general agreement with experiments by Ribbeck and Clausen (Clausen and Ribbeck, 2007) who measured total microtubule mass after RanQ69L addition. Addition of TPX2 as well as RanQ69L further accelerated the rate of formation as well as the number of microtubules compared to RanQ69L alone.
Figure 2. Quantifying the effect of RanQ69L and TPX2 on microtubule nucleation.
The number of EB1 spots is counted for each time frame and experimental condition, and plotted against time. Because each microtubule, even when branched, is marked by a single EB1 spot at its growing plus tip, counting the number of EB1 spots directly corresponds to the number of microtubules per frame and field of view (82.2 × 82.2 μm). Data from a control extract in which only vanadate was added or experiments in which further additions of RanQ69L, TPX2, or RanQ69L plus TPX2 were made.
We next examined the angles of daughter filaments that emerge from the mother microtubule; a branch angle of zero degrees is defined as a parallel growth and 180° as antiparallel growth (Fig. 3A). The branch angle was shallow in most instances, with the daughter microtubule growing parallel or nearly parallel to the template microtubule (<10° for 52% of branching events) (Fig. 3A and Movies S1, S2 and S3). Strikingly, the daughter filament virtually always grows toward the plus end of the template microtubule (94% of the branch angles of daughter MT are <90° with RanQ69L addition (Fig. 3A and Movies S1, S2 and S3)), thus replicating the polarity of parent microtubules in the emerging network.
Figure 3. Properties of branching microtubule nucleation in a RanQ69L-treated extract.
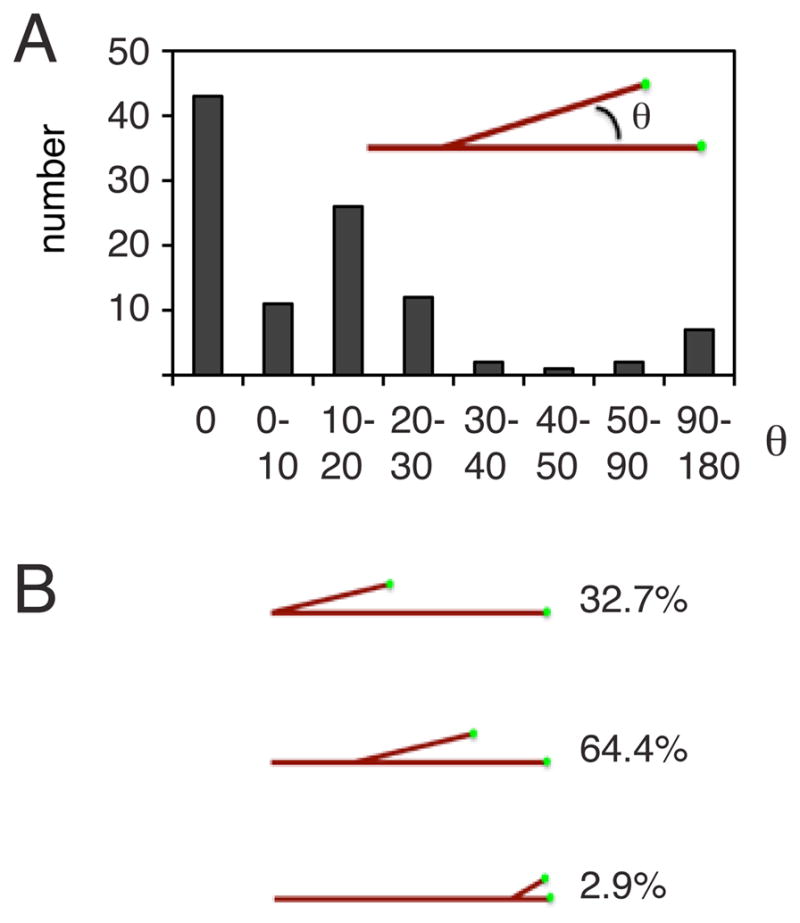
(A) Quantification of branch angles between the mother microtubule and the newly growing daughter microtubule. A 0 degree angle corresponds to growth of a new microtubule parallel to the mother microtubule, whereas a 180 degree angle signifies antiparallel growth. The polarity of the mother microtubule is preserved if the branch angle is smaller than 90°. 89% of the branches are <30°. (B) Quantification of branch position along the template microtubule. The majority of branches occur along the middle of the microtubule, about a third originate at the microtubule minus end and more rarely the EB1-enriched microtubule plus tip branches.
Microtubule nucleation occurred predominantly along the length of the original microtubule (~60%) and at the very distal minus end (~30% of events, Fig. 3B). Many nucleation events along the length of the microtubule occurred at the same site (within the resolution of the microscope) of a previous nucleation event (~33%). In rare cases, a growing EB1 comet appeared to split apart, thereby generating two new microtubules from a growing plus end.
Molecular factors required for branching microtubule nucleation
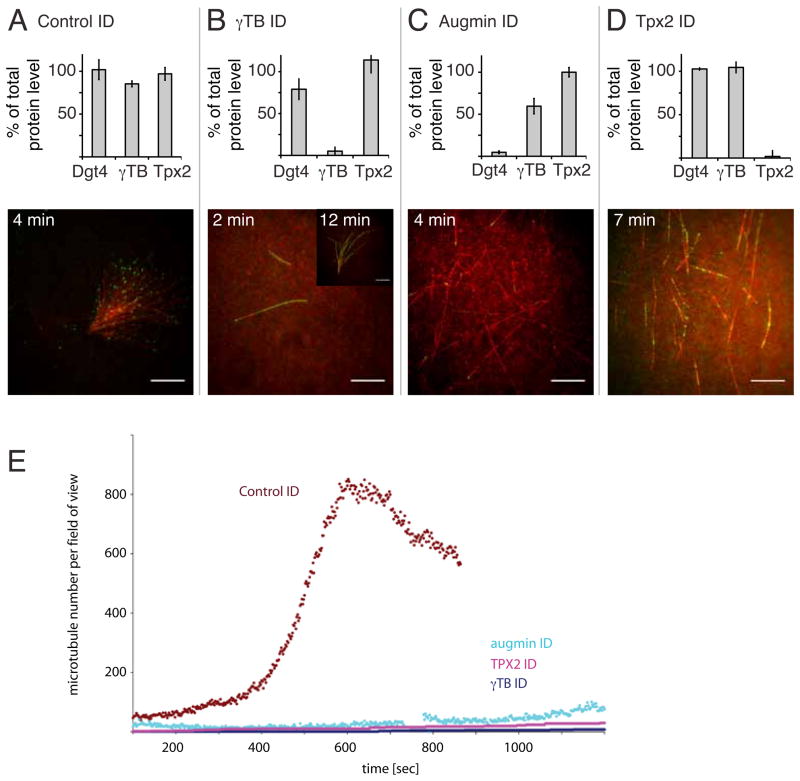
Using the robust branching microtubule nucleation assay in the presence of both RanQ69L and TPX2, we tested for the involvement of additional factors by immunodepletion. Formation of fan-shaped, branching microtubule structures that originated by branching microtubule nucleation) was not affected by a control immunodepletion with nonspecific IgG antibodies (Fig. 4A, Movie S6; 1.42 ± 0.56 fans (defined as a branching cluster of >5 MT) per 54 × 54 μm field of view; mean and sem of 2 independent experiments with ~50 fields each). However, after immunodepletion of γ-TB (~95%; Fig. S2B, (Wiese and Zheng, 2000)), very few microtubules formed within 7 min (Fig. 4B, Movie S6) and those that could be observed grew very long. The formation of these few microtubules might be due to the activity of residual γ-TuRC or some minor alternative pathway. Although rare on the coverslip, fans composed of few and unusually long microtubules were observed in γ-TB-depleted extract (insert, Fig. 4B; Movie S6)(0.04 ± 0.02 fans/field of view; mean and sem of 2 independent experiments with ~50 fields each). These data indicate that γ-TuRC is required for the primary nucleation pathway.
Figure 4. Molecular factors required for branching microtubule nucleation in the presence of RanQ69L and TPX2.
(A) A control immunodepletion of the extract with total IgG fraction antibodies still produced fan-shaped structures composed of branching microtubule and did not significantly affect levels of augmin (Dgt4 subunit), γ-TB and TPX2 levels as quantified in a Western blot (bottom, error bars represent s.d. of three independent experiments with different extracts). (B) Immunodepletion of γ-TB (bottom) almost extinguished microtubule nucleation; most fields had no or a few microtubules; after longer times (7 min), rare examples of fan-shaped structures with few, but long microtubules could be found on the coverslip (see insert). (C) Immunodepletion of the augmin Dgt4 subunit, which depletes the entire augmin complex and one third of γ-TB (bottom diagram), abolished branching microtubule nucleation, but still allowed for microtubule growth. (D) Similarly, immunodepletion of TPX2 abolished branching microtubule nucleation, but still allowed for microtubule growth. Scale bars, 10 μm. See The time shown indicates the approximate time of recording after adding the extracts to the flow cell. Movie S6 for sequences of these immunodepletions. See also Figure S2. (E) Quantification of microtubule number over time for different immunodepletions. Reactions were made in the presence of RanQ69L plus TPX2 (except for the TPX2 depletion, in which case only RanQ69L was added).
We next investigated the role of augmin, a protein complex shown to target γ-TB to pre-existing microtubules (Goshima et al., 2008; Goshima et al., 2007). Unlike γ–TB depletion, microtubule nucleation and growth still occurred after augmin immunodepletion (Fig. S2B), although after a delay compared to the control immunodepletion (Fig. 4C, Movie S6). Strikingly, branching events and the formation of fan-shaped structures were virtually abolished (no fans observed in three independent experiments); instead individual microtubules formed gradually. This effect is not due to partial (~30%) γ-TB depletion that accompanies augmin immunodepletion (Petry et al., 2011). Similar results were obtained after TPX2 immunodepletion (Fig. S2A); individual microtubules appeared on the glass surface after a delay, but branching microtubule nucleation was not observed and fan-shaped structures did not form (Fig. 4D, Movie S6; no fans observed in three independent experiments). Collectively these results reveal that γ-TB is required for virtually all nucleation activity and that both augmin and TPX2 have an essential role in promoting branching microtubule nucleation.
We next examined the effects of augmin, TPX2, or γ-TB depletion after RanGTP/TPX2 addition on the kinetics of microtubule nucleation. Strikingly, in all cases, with the γ-TB depletion being the most severe, the initial sigmoidal increase in microtubule number virtually disappeared and only a slow increase in microtubule number was observed over the first 20 min of the reaction (Fig. 4E). This result, combined with the observation that both augmin and TPX2 are required for the appearance of microtubule branches (Fig. 4C and 4D), suggests that branching microtubule nucleation is the major mechanism responsible for the acceleration and amplification of microtubule numbers that occurs when RanGTP is added to the extract.
TPX2 interactions
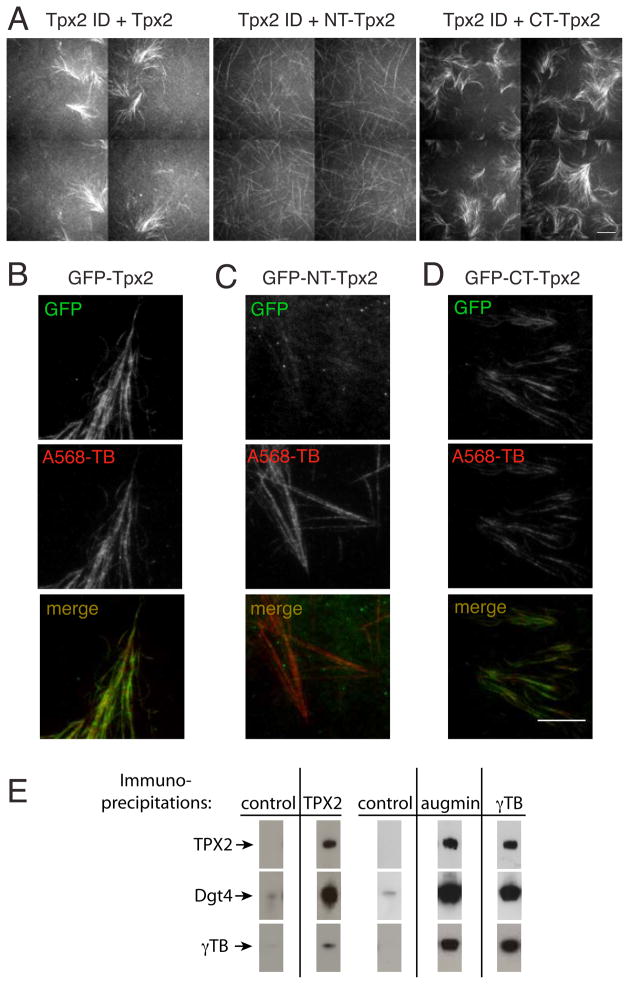
TPX2 is a multi-domain protein that has been implicated in many activities during spindle assembly. The N-terminal portion of TPX2 binds to and activates Aurora A kinase (Tsai et al., 2003); its C-terminus has been implicated in direct microtubule binding and interactions with the kinesins Eg5 and Xklp2 (Eckerdt et al., 2008; Wittmann et al., 1998). We prepared recombinant N- and C-terminal TPX2 fragments (NT- and CT-TPX2) and asked whether they could restore branching microtubule nucleation after depleting endogenous TPX2. Add-back of NT-TPX2 did not rescue branching microtubule nucleation (Fig. 5A, Movie S7; no fans in two independent experiments); the phenotype was indistinguishable from TPX2-depletion alone (Fig. 4D). In contrast, add-back of CT-TPX2 restored branching microtubule nucleation (Fig. 5A, Movie S7) (1.73 ± 1.3 fans/field of view; mean and sem of 2 independent experiments with ~50 fields each), as did full-length TPX2 (0.77 ± 0.14 fans/field of view; mean and sem of 2 independent experiments with ~50 fields each). Using GFP-fusion proteins, we found that full length TPX2 and CT-TPX2, but not NT-TPX2, were recruited to the fan-shaped microtubule networks (Fig. 5B–D). Thus, TPX2 and CT-TPX2 bound to microtubules where branching nucleation occurred, although they did not necessarily localize exclusively to sites of branching.
Figure 5. Rescue of branching microtubule nucleation by addition of recombinant TPX2 to TPX2-depleted Xenopus extracts.
(A) Addition of purified TPX2 or CT-TPX2 to extract depleted of TPX2 (see Fig. 3D) could rescue branching microtubule nucleation and the formation of branched fan-like structures. In contrast, add-back of NT-TPX2 displayed the same phenotype as TPX2 immunodepletion (see Fig. 3D). For each experimental condition a montage of four adjacent fields is displayed, which was collected with the slide explorer function of the MicroManager software (Edelstein et al., 2010). Scale bar, 10 μm. See Movie S7. (B) Localization of full-length GFP-TPX2 (C) Localization of the N-terminus of GFP-TPX2 (GFP-NT-TPX2) (D) Localization of the C-terminus of GFP-TPX2 (GFP-CT-TPX2). The full-length and GFP-CT-TPX2 constructs binds along microtubules of the fan-like structures, whereas the N-terminus of GFP-TPX2 can hardly be detected. Scale bar, 10 μm. E) Immunoprecipitations using antibodies specific against TPX2, augmin, and γ-TB, and control antibodies (total IgG fraction antibodies). The same antibodies were used in the immunoblot as detection reagents. TPX2, augmin, and γ-TB interact with each other. The two controls reflect separate immunoprecipitation experiments.
Augmin and γ-TB were previously shown to interact by immunoprecipitation experiments (Petry et al., 2011; Uehara et al., 2009; Zhu et al., 2008). We next tested whether endogenous TPX2 in the extracts interacts with augmin or γ-TB. When TPX2 was immunoprecipitated on beads, augmin and γ-TB were found to co-immunoprecipitate in a specific manner compared to control beads (Fig. 5E). Similarly, immunoprecipitation of augmin and γ-TB resulted in a specific signal for TPX2 on these beads. These experiments do not reveal the stoichiometry of interactions between these proteins nor whether the interactions are direct or mediated through other proteins. However, the immunoprecipitation experiments raise the possibility that these proteins may cooperate in a complex that promotes microtubule nucleation.
Discussion
In this study, we have developed a new assay for visualizing branching microtubule nucleation in meiotic Xenopus egg extracts and characterized its properties. This is the first time that branching microtubule nucleation has been visualized in an animal cell system. Daughter microtubules grow in a low branch angle and thus preserve polarity, making branching microtubule nucleation an ideal mechanism for generating parallel microtubule structures, such as the mitotic spindle. We found that RanGTP and its downstream target TPX2 stimulate branching microtubule nucleation and that augmin, γ-TB and TPX2 are essential for this process.
The branching nucleation mechanism catalyzes microtubule production and preserves polarity of microtubule structures in Xenopus extracts
In Xenopus egg extracts, the increase in microtubule mass over time following the addition of chromatin beads (Dinarina et al., 2009; Groen et al., 2009) or RanQ69L (Clausen and Ribbeck, 2007) demonstrates nonlinear behavior characterized by an initial lag, a steeply rising phase, and then a plateau. Clausen and Ribbeck (Clausen and Ribbeck, 2007) showed that addition of seed microtubules shifts this sigmoidal curve to earlier time points. They suggested that microtubules induce their own production in an autocatalytic manner, perhaps through a phenomenon of microtubule-dependent microtubule nucleation (Mahoney et al., 2006). In this study, we have directly measured microtubule number by counting EB1 spots and observed a similar sigmoidal increase over time with RanQ69L and a further enhancement in the rate by addition of TPX2.
Strikingly, immunodepletion of augmin abolishes the sigmoidal increase of microtubule number as a function of time after RanQ69L/TPX2 addition and instead results in a slow linear increase. This quantitative analysis of microtubule number is consistent with prior results showing a delay in the formation of Xenopus acentrosomal spindles and RanQ69L-mediated asters after augmin depletion (Petry et al., 2011). Furthermore, we have found that branching microtubule nucleation is absent after augmin immunodepletion. These results suggest that branching microtubule nucleation is the primary mechanism responsible for the rapid, sigmoidal increase in microtubule numbers observed after RanQ69L and TPX2 addition. This conclusion is predicated on augmin only enhancing microtubule numbers through a branching nucleation mechanism, an assumption supported by current data showing a role for augmin in the recruitment γ-TuRC to the side of a pre-existing microtubule. However, one cannot exclude other nucleation promoting activities for augmin in solution.
Since daughter microtubules virtually always grow in the same direction as the mother microtubule, branching microtubule nucleation is ideal for quickly generating parallel microtubule structures in the cell. This feature distinguishes animal cell microtubule branching from that in fission yeast, where branching nucleation generates microtubules of opposite polarity and hence anti-parallel microtubule structures (Janson et al., 2005). This result, combined with the fact that augmin homologues have not been found in yeast, suggests that branching nucleation in yeast and metazoans occur by fundamentally different mechanisms. In plants, augmin has recently been shown to be important for microtubule production in the spindle and phragmoplasts of plants (Ho et al., 2011; Hotta et al., 2012; Nakaoka et al., 2012). Branching microtubule nucleation also has been observed in plants, although not yet identified as an activity of augmin (Chan et al., 2009; Murata et al., 2005). In Arabidopsis, the daughter-mother branch angles are more broadly distributed (0–60°) (Chan et al., 2009; Murata et al., 2005) than what we describe in this study (89% between 0–30°).
Molecular players of branching microtubule nucleation
Since augmin’s discovery, it has been speculated that it may be involved in a branching microtubule nucleation mechanism (Goshima et al., 2008; Goshima et al., 2007). This hypothesis was based circumstantially on the findings that i) augmin itself localizes to spindle microtubules, ii) that it is needed to recruit γ-TB to the spindle microtubules, and iii) that its depletion by RNAi decreases microtubule density within the spindle (Goshima et al., 2008; Goshima et al., 2007; Hotta et al., 2012; Lawo et al., 2009; Nakaoka et al., 2012; Petry et al., 2011; Uehara and Goshima, 2010; Uehara et al., 2009; Zhu et al., 2008). Here, using single microtubule imaging, we provide direct evidence for this hypothesis by showing that branching microtubule nucleation takes place in meiotic Xenopus egg extracts and that augmin depletion abolishes this reaction.
While the connection between augmin and branching nucleation is not necessarily surprising based upon past work, the involvement of TPX2 in this reaction was not necessarily expected. Previous studies have shown that TPX2 is a key factor that promotes microtubule assembly in Xenopus egg extracts and is regulated by Ran (Gruss et al., 2001). TPX2 also was found to promote the assembly of microtubules from pure tubulin in solution vitro (Brunet et al., 2004; Schatz et al., 2003). Here, we show that TPX2 immunodepletion inhibits branching microtubule nucleation, connecting TPX2 with the augmin- and γ-TuRC-mediated nucleation reaction for the first time. TPX2 interacts with both augmin and γ-TuRC by immunoprecipitation, suggesting the possibility of the assembly of a larger branching nucleation complex. From these results, one might expect augmin, TPX2 and γ-TuRC to be localized at branch points. However, our current results with antibodies to these proteins reveal general staining throughout the microtubule “fan”, making it difficult to discern specific localization at branches (unpublished results). Further insight into the molecular mechanism of how augmin, TPX2 and γ-TuRC give rise to branching microtubule nucleation will require in vitro reconstitution of this nucleation process with purified proteins.
The Ran pathway, augmin and microtubule branching nucleation
Our recent study showed that augmin depletion inhibited RanGTP-mediated microtubule aster formation in Xenopus extracts (Petry et al., 2011), but the connection to RanGTP was not previously evident. The present studies show directly that branching microtubule nucleation is stimulated by RanGTP and solidifies a connection between upstream formation of RanGTP, created by the chromosome-bound guanine-nucleotide exchange factor RCC1, and the downstream microtubule nucleation by augmin and γ-TuRC. Most studies to date propose that RanGTP stimulates microtubule nucleation in the vicinity of chromosomes or as a gradient decaying from chromatin, rather than occurring throughout the body of the spindle (Caudron et al., 2005; Kalab et al., 2006; Clausen and Ribbeck, 2007). However, microtubule nucleation (as evidenced by the appearance of GFP-EB1 comets) occurs throughout the spindle, even at acentrosomal poles located at a considerable distance from the chromosomes (Mahoney et al, 2006; Brugues et al., 2012). Furthermore, models in which RanGTP stimulates soluble microtubule nucleators would be expected to create randomly oriented microtubules, which would require continual sorting by microtubule motors (Walczak et al., 1998) to maintain the polarity of microtubules in the spindle. These aspects of the Ran-mediated microtubule nucleation become less puzzling if one of its downstream microtubule nucleators is located on the microtubule itself, using the information of pre-existing microtubules to define the polarity of new microtubules. Thus, the actions of microtubule motors and branching nucleation can synergize to create and maintain the polarity of microtubules in the spindle. However, it remains an open question of how the combined actions of RanGTP, the upstream activator emanating from the chromosomes, and augmin/γ-TuRC, downstream effectors located on microtubules throughout the spindle (Goshima et al., 2008; Lawo et al., 2009; Uehara et al., 2009; Zhu et al., 2008), regulate branching microtubule nucleation spatially and temporally during spindle assembly.
Role of branching microtubule nucleation in spindle assembly
Branching microtubule nucleation may help to explain several unresolved aspects of meiotic and mitotic spindles, including their rapid assembly, the maintenance of local structure and polarity despite rapid microtubule turnover, and the sharp boundaries at the edge of spindle. Computational studies suggest that the two originally proposed pathways of centrosome- and chromatin-based microtubule nucleation are insufficient to explain these features of spindle assembly (Clausen and Ribbeck, 2007; Loughlin et al., 2010; Needleman et al., 2010). Our observations of branching MT nucleation, specifically the low branch angles and the contribution of augmin/TPX2-mediated branching to the sigmoidal increase in microtubule numbers, are consistent with this nucleation mechanism contributing to (i) efficient amplification (during assembly) and maintenance (at steady state) of microtubule density in the spindle, (ii) preservation of local polarity throughout the spindle, and (iii) the creation of edges through strong positive feedback of microtubule density. These microscopic features are also consistent with the gross perturbations that occur in the spindle in augmin-depleted cells or Xenopus extracts, which include a decrease in spindle microtubule density and poor spindle shape.
Branching microtubule nucleation is also well suited to build and maintain parallel, high-density microtubule bundles, such as kinetochore fibers. Consistent with this idea, augmin depletion reduces kinetochore fiber formation and stability (Bucciarelli et al., 2009; Goshima et al., 2008; Lawo et al., 2009; Uehara et al., 2009), augmin localizes to kinetochore fibers in plants (Hotta et al., 2012) and TPX2 localizes preferentially to kinetochore fibers in human cells (Bird and Hyman, 2008). The augmin subunit Dgt6 also interacts with the kinetochore protein Ndc80 (Bucciarelli et al., 2009) and addition of excess importin β or depletion of TPX2 blocks microtubule formation from kinetochores (Tulu et al., 2006). Thus, augmin and TPX2 might be involved in promoting microtubule nucleation both at kinetochores as well as along kinetochore fibers.
Experimental Procedures
Proteins, antibodies and Xenopus egg extract generation
Purification of GFP-EB1 and production of the antibodies against Xenopus laevis Dgt4 and γ-TB were described recently (Petry et al., 2011). The anti-TPX2 antibody (Groen et al., 2009), and GST-tagged TPX2 (Groen et al., 2004) were prepared as described earlier. GFP-tagged full-length, N-terminal and C-terminal TPX2 constructs (Brunet et al., 2004) were bacterially expressed and purified by affinity-chromatography with Ni-NTA (Qiagen). RanQ69L was purified in the same way, but additionally loaded with GTP as described (Weis et al., 1996). The CC1 construct (P150-CC1) was expressed and purified as described previously (King et al., 2003). CSF extracts were prepared from Xenopus laevis oocytes according to (Murray and Kirschner, 1989) and either used immediately or subjected to immunodepletions (Petry et al., 2011).
Assay set-up, microscopy and image analysis
Untreated or immunodepleted CSF extracts were supplemented with 0.86 μM Alexa568-labeled bovine brain tubulin and 0.8 μM GFP-EB1. Depending on the experiment, 0.5 mM vanadate (sodium orthovanadate), 11 μM RanQ69L, or 1 μM TPX2 (or the NT or CT TPX2 construct) were further added. The CC1 construct was substituted for vanadate at a final concentration of 0.125mg/ml. Each component was added from 20x stock solution in CSF-XB buffer to minimize dilution. For control experiments, an equivalent volume of CSF-XB buffer was used. To start the experiment, a 5 μl reaction mixture (previously kept on ice) was pipetted into a flow cell composed of a glass slide, two stripes of double-stick tape, and a glass coverslip, and examined with total internal reflection fluorescence (TIRF) microscopy. Microtubule nucleation and the formation of branched fan-like structures can be observed several minutes after pipetting the reaction mix into the flow cell. This delay time varies slightly with each extract and increases with the dilution of extract in the reaction mix.
For the experiment with a pig brain template microtubule, glass coverslips were first passivated with dichlorodymethylsilane (Gell et al., 2010). The glass surface was coated with an anti-biotin antibody and blocked with κ-casein before attaching biotin- and Cy5-labeled microtubules, which were stabilized by GMPCPP. After a final wash of the flow cell with CSF-XB, CSF extract was added and imaging started.
Imaging was performed with TIRF microscopy on a Nikon Ti or Eclipse TE200-E microscope equipped with an Andor Ixon EM CCD camera, a 100x, 1.49 NA TIRF objective and Micro-Manager imaging software (Edelstein et al., 2010). Dual and triple color images were acquired every 2 sec at room temperature. ImageJ (Wayne Rasband, NIH) was used for quantitating branch angles between the daughter and mother microtubule. An angle of 0 degrees corresponds to parallel growth.
For quantification of microtubule numbers, the GFP-EB1 spots of an image sequence were sharpened by applying a Gaussian Blur function with a low (1) and high (5) radius of decay, followed by subtraction of the latter image sequence from the first one. Next, a threshold was set to eliminate background noise before counting particles with the Analyze Particles function in ImageJ (Wayne Rasband, NIH).
Supplementary Material
01
02
03
04
05
06
07
08
Highlights.
- A new assay was developed for visualizing MT nucleation from the sides of existing MTs.
- Newly nucleated daughter MTs grow with the same polarity as the mother MT.
- Branching MT nucleation is stimulated by RanGTP and its effector TPX2.
- Augmin, shown earlier to increase spindle MT density, is required for branching MT nucleation.
Acknowledgments
We thank Steve Ross and Nikon, Inc. for generously providing a microscope to conduct initial studies at the MBL, Woods Hole. We thank Rebecca Heald, Kara Helmke and Jeremy Wilbur (UC Berkeley) for generous help with Xenopus egg extracts, providing vectors, and suggestions. We also thank Chris Field and Ani Nguyen (Harvard Medical School) for help with Xenopus egg extracts, Jim and Cathy Galbraith (NIH) and Nico Stuurman (UCSF) for microscopy assistance. This work was supported by an NIH grants GM38499 to RDV and GM39565 to TJM, an Evans Foundation Fellowship at MBL to RDV and TJM, and a Life Science Research Foundation fellowship sponsored by HHMI and a K99 NIH grant to S.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Athale CA, Dinarina A, Mora-Coral M, Pugieux C, Nedelec F, Karsenti E. Regulation of microtubule dynamics by reaction cascades around chromosomes. Science. 2008;322:1243–1247. doi: 10.1126/science.1161820. [DOI] [PubMed] [Google Scholar]
- Bird AW, Hyman AA. Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J Cell Biol. 2008;182:289–300. doi: 10.1083/jcb.200802005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- Brinkley BR. Microtubule organizing centers. Annu Rev Cell Biol. 1985;1:145–172. doi: 10.1146/annurev.cb.01.110185.001045. [DOI] [PubMed] [Google Scholar]
- Brugues J, Nuzzo V, Mazur E, Needleman DJ. Nucleation and transport organize microtubules in metaphase spindles. Cell. 2012;149:554–564. doi: 10.1016/j.cell.2012.03.027. [DOI] [PubMed] [Google Scholar]
- Brunet S, Sardon T, Zimmerman T, Wittmann T, Pepperkok R, Karsenti E, Vernos I. Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol Biol Cell. 2004;15:5318–5328. doi: 10.1091/mbc.E04-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucciarelli E, Pellacani C, Naim V, Palena A, Gatti M, Somma MP. Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and gamma-tubulin to promote kinetochore-driven MT formation. Curr Biol. 2009;19:1839–1845. doi: 10.1016/j.cub.2009.09.043. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Caudron M, Bunt G, Bastiaens P, Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- Chan J, Sambade A, Calder G, Lloyd C. Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell. 2009;21:2298–2306. doi: 10.1105/tpc.109.069716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T, Ribbeck K. Self-organization of anastral spindles by synergy of dynamic instability, autocatalytic microtubule production, and a spatial signaling gradient. PLoS ONE. 2007;2:e244. doi: 10.1371/journal.pone.0000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarina A, Pugieux C, Corral MM, Loose M, Spatz J, Karsenti E, Nedelec F. Chromatin shapes the mitotic spindle. Cell. 2009;138:502–513. doi: 10.1016/j.cell.2009.05.027. [DOI] [PubMed] [Google Scholar]
- Dumont J, Desai A. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol. 2012;22:241–249. doi: 10.1016/j.tcb.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerdt F, Eyers PA, Lewellyn AL, Prigent C, Maller JL. Spindle pole regulation by a discrete Eg5-interacting domain in TPX2. Curr Biol. 2008;18:519–525. doi: 10.1016/j.cub.2008.02.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using microManager. In: Ausubel Frederick M, et al., editors. Curr Protocols Mol Biol. Chapter 14. 2010. pp. 20.2–20.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell C, Bormuth V, Brouhard GJ, Cohen DN, Diez S, Friel CT, Helenius J, Nitzsche B, Petzold H, Ribbe J, et al. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods in cell biology. 2010;95:221–245. doi: 10.1016/S0091-679X(10)95013-9. [DOI] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenan G, Brangwynne CP, Jaensch S, Gharakhani J, Julicher F, Hyman AA. Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr Biol. 2010;20:353–358. doi: 10.1016/j.cub.2009.12.050. [DOI] [PubMed] [Google Scholar]
- Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ, Ohi R. XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr Biol. 2004;14:1801–1811. doi: 10.1016/j.cub.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Groen AC, Maresca TJ, Gatlin JC, Salmon ED, Mitchison TJ. Functional Overlap of Microtubule Assembly Factors in Chromatin-Promoted Spindle Assembly. Mol Biol Cell. 2009;20:2766–2773. doi: 10.1091/mbc.E09-01-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Ho CM, Hotta T, Kong Z, Zeng CJ, Sun J, Lee YR, Liu B. Augmin plays a critical role in organizing the spindle and phragmoplast microtubule arrays in Arabidopsis. Plant Cell. 2011;23:2606–2618. doi: 10.1105/tpc.111.086892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta T, Kong Z, Ho CM, Zeng CJ, Horio T, Fong S, Vuong T, Lee YR, Liu B. Characterization of the Arabidopsis augmin complex uncovers its critical function in the assembly of the acentrosomal spindle and phragmoplast microtubule arrays. Plant Cell. 2012;24:1494–1509. doi: 10.1105/tpc.112.096610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson ME, Setty TG, Paoletti A, Tran PT. Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J Cell Biol. 2005;169:297–308. doi: 10.1083/jcb.200410119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Karsenti E, Newport J, Hubble R, Kirschner M. Interconversion of metaphase and interphase microtubule arrays, as studied by the injection of centrosomes and nuclei into Xenopus eggs. J Cell Biol. 1984;98:1730–1745. doi: 10.1083/jcb.98.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol. 2000;2:352–357. doi: 10.1038/35014045. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer TA, Sillje HH, Korner R, Gruss OJ, Meraldi P, Nigg EA. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JR, Hegemann B, et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- Loughlin R, Heald R, Nedelec F. A computational model predicts Xenopus meiotic spindle organization. J Cell Biol. 2010;191:1239–1249. doi: 10.1083/jcb.201006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney NM, Goshima G, Douglass AD, Vale RD. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr Biol. 2006;16:564–569. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, Horio T, Hasebe M. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat Cell Biol. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- Nakaoka Y, Miki T, Fujioka R, Uehara R, Tomioka A, Obuse C, Kubo M, Hiwatashi Y, Goshima G. An inducible RNA interference system in Physcomitrella patens reveals a dominant role of augmin in phragmoplast microtubule generation. Plant Cell. 2012;24:1478–1493. doi: 10.1105/tpc.112.098509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman DJ, Groen A, Ohi R, Maresca T, Mirny L, Mitchison T. Fast microtubule dynamics in meiotic spindles measured by single molecule imaging: evidence that the spindle environment does not stabilize microtubules. Mol Biol Cell. 2010;21:323–333. doi: 10.1091/mbc.E09-09-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Petry S, Pugieux C, Nedelec FJ, Vale RD. Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc Natl Acad Sci USA. 2011;108:14473–14478. doi: 10.1073/pnas.1110412108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Poleward microtubule flux mitotic spindles assembled in vitro. J Cell Biol. 1991;112:941–954. doi: 10.1083/jcb.112.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz CA, Santarella R, Hoenger A, Karsenti E, Mattaj IW, Gruss OJ, Carazo-Salas RE. Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 2003;22:2060–2070. doi: 10.1093/emboj/cdg195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Salmon ED, Mitchison TJ. Microtubule plus-end dynamics in Xenopus egg extract spindles. Mol Biol Cell. 2004;15:1776–1784. doi: 10.1091/mbc.E03-11-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MY, Wiese C, Cao K, Martin O, Donovan P, Ruderman J, Prigent C, Zheng Y. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat Cell Biol. 2003;5:242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- Tulu US, Fagerstrom C, Ferenz NP, Wadsworth P. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr Biol. 2006;16:536–541. doi: 10.1016/j.cub.2006.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Goshima G. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J Cell Biol. 2010;191:259–267. doi: 10.1083/jcb.201004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Nozawa RS, Tomioka A, Petry S, Vale RD, Obuse C, Goshima G. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc Natl Acad Sci USA. 2009;106:6998–7003. doi: 10.1073/pnas.0901587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- Weis K, Dingwall C, Lamond AI. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J. 1996;15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- Wiese C, Zheng YX. Microtubule nucleation: gamma-tubulin and beyond. J Cell Sci. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
- Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Boleti H, Antony C, Karsenti E, Vernos I. Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J Cell Biol. 1998;143:673–685. doi: 10.1083/jcb.143.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dawe RK. Mechanisms of plant spindle formation. Chromosome Research. 2011;19:335–344. doi: 10.1007/s10577-011-9190-y. [DOI] [PubMed] [Google Scholar]
- Zhu H, Coppinger JA, Jang CY, Yates JR, 3rd, Fang G. FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J Cell Biol. 2008;183:835–848. doi: 10.1083/jcb.200807046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05
06
07
08