The importance of phase information for human genomics (original) (raw)
. Author manuscript; available in PMC: 2013 Aug 27.
Published in final edited form as: Nat Rev Genet. 2011 Feb 8;12(3):215–223. doi: 10.1038/nrg2950
Abstract
Contemporary sequencing studies often ignore the diploid nature of the human genome because they do not routinely separate or ‘phase’ maternally and paternally derived sequence information. However, many findings — both from recent studies and in the more established medical genetics literature — indicate that relationships between human DNA sequence and phenotype, including disease, can be more fully understood with phase information. Thus, the existing technological impediments to obtaining phase information must be overcome if human genomics is to reach its full potential.
Advances in DNA-sequencing technologies have made it possible to efficiently characterize large segments of, if not entire, individual human genomes1, 2, 3, 4. Sequencing the genomes of members of the same family4, from individuals with and without a particular disease5, or from individuals sampled randomly from the population6, can lead to insight into the role of both common and rare DNA sequence variants in mediating phenotypic expression. However, most studies of this kind typically involve sequencing DNA samples that contain both the maternally and the paternally derived DNA associated with the homologous chromosomes inherited by an individual. As such, they essentially ignore the phase of the DNA in those samples — that is, they ignore the unique nucleotide content of the two homologous chromosomes an individual possesses, referred to as an individual’s ‘diplotype’. Human genome-related initiatives, such as the International HapMap Project and the 1000 Genomes Project, have considered the importance of haplotyping. However, this is usually in the service of assessing, through linkage-disequilibrium measures, the likelihood that variants at one genomic position indicate the presence of variants at neighbouring positions. Rarely does contemporary consideration of phase information concern the molecular physiological consequences of having variants uniquely distributed across two homologous chromosomal copies of a genomic region7.
The dearth of phased human genomic data is primarily due to the computational complexity associated with, and the lack of cost-effective approaches for, obtaining phase information. Well-established phenomena such as compound heterozygosity in monogenic disorders support the importance of phase information for relating genotype to phenotype. In addition, recent studies have described settings in which the characterization of the specific nucleotides on each homologous copy of a gene or genomic region inherited by an individual is essential for understanding phenotypic expression4, 8, 9, 10, 11. Here, we discuss these studies and consider specific instances in which the specific set of variants on each homologous chromosome contributes to phenotypic expression and disease states. We also briefly describe other settings in which phase information is important for human genomics research. We provide an overview of current methods for obtaining phase information, and discuss their limitations and prospects for future improvement. We also coin the term ‘diplomics’ to refer to scientific investigations that leverage phase information in order to understand how molecular and clinical phenotypes are influenced by unique diplotypes. We ultimately argue that diplomic investigations will be key to the design and conduct of future functional genomic studies, as well as large-scale human DNA-sequencing initiatives.
Diplotype is important for function
To understand the importance of phase information in human sequencing studies, it is necessary to understand the settings in which the balance of _cis_- and _trans_-acting variants on the two homologous copies of a genomic region affect phenotypic expression (Fig. 1). A number of recent studies have used high-throughput DNA sequencing to investigate how nucleotide variation affects gene function in a way that depends on which chromosome
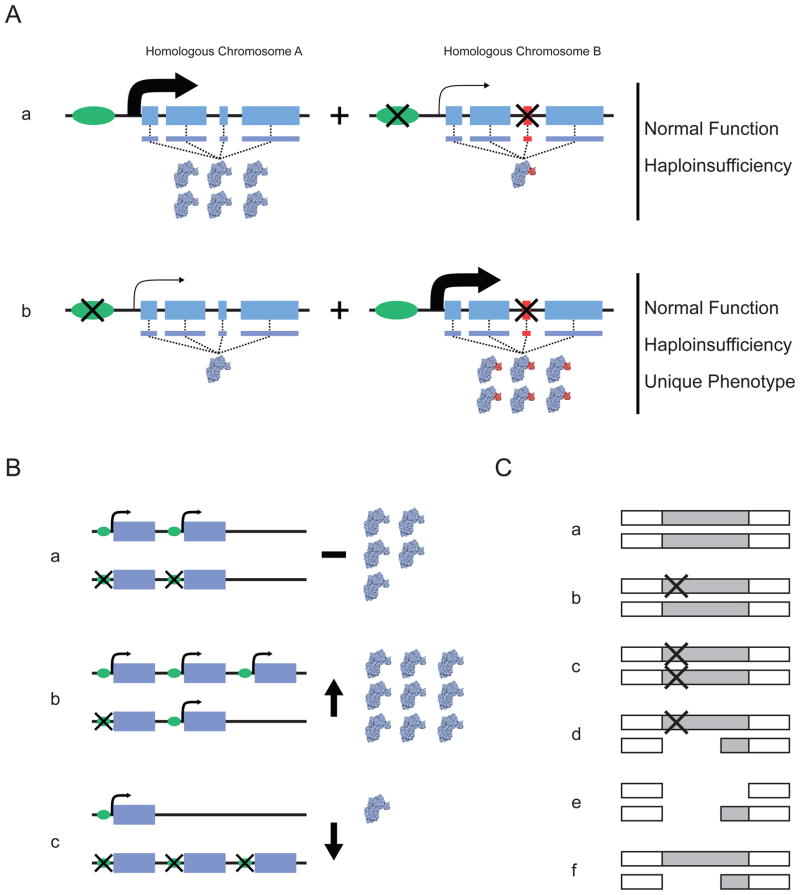
Figure 1. The distribution of variants between homologous chromosomes can affect gene function.
A | Distribution of variants that affect regulation and protein function, showing the two homologous gene segments in a single diploid individual. Aa | In this case, the leftmost homologue does not contain variation that influences either the expression or the structure of the encoded protein. By contrast, the rightmost homologue contains sequence variation in the promoter that reduces overall expression of the gene and exonic sequence variation that upsets the amino-acid sequence of the encoded protein. Ab | Here, the variants in the promoter and exonic sequence are distributed between different homologues. The combination of these homologues in a single individual can lead to haploinsufficiency if the homologue that does not have a functional variant cannot compensate for the affected homologue. If it can compensate, the overall functioning of the gene could be normal, owing to both the downregulation of the aberrant protein and the normal expression of the wild-type protein. B | Potential functional effects of haplotypes involving structural variants. Scenarios are shown involving copy-number variants and point mutations in a diploid setting. The possibilities depicted in parts Bb and Bc reflect increased and decreased overall gene expression, respectively, relative to that in Ba. C | Unmasking of deleterious mutations through gene deletion. A genomic region is shown that harbours a gene that is often either partially or completely deleted and that also harbours functionally relevant point mutations. Ca | Neither homologous copy of the gene harbours a variant. Cb | One of the gene homologues carries a point mutation. Cc | Both gene homologues carry a point mutation. Cd | One of the gene homologues carries a deletion and the other carries a point mutation. Ce | Both of the gene homologues carry a deletion. Cf | One of the gene homologues carries a deletion. Each situation could produce a different phenotype; for example, in part Cd the deletion depicted could unmask the deleterious effect of the point mutation on the other chromosome.
Widespread allele-specific expression
The ability of a cell to selectively express a gene on a single chromosome while the gene on the homologous chromosome is silenced is a well-characterized phenomenon in diploid cells. This effect can be caused by, but is not necessarily limited to, nucleotide variation or methylation at the locus that regulates or harbours the affected gene. Recent studies have indicated that such allele-specific expression (ASE) is widespread in humans. Two groups recently used RNA sequencing to study how _cis_-acting sequence variation influences gene expression10, 11. Both groups showed that 1–5% of human genes are influenced by _cis_-acting DNA sequence variants (known as expression QTLs, or eQTLs) in the contexts that they tested. Most heterozygous _cis_-acting eQTLs resulted in one copy of the gene being expressed at a higher level than its homologous copy — hence exhibiting ASE. There are a number of possible biological mechanisms responsible for ASE. Kasowaki et al.8, for example, showed that the binding strengths of two transcription factors (TFs) exhibit wide variation at ~25% of specific TF target sequences across different individuals. Differences in binding strength across individuals were frequently associated with the existence of genetic variants in these binding regions. Such differences in binding strength were not only shown to be correlated with differences in the expression levels of genes associated with the TF target sites, but also to have clear segregation in families — therefore exhibiting heritability — thus confirming the genetic origins of the variation in gene expression levels8, 15.
Epigenetic changes in a genomic region can also influence gene expression in a chromosome- or allele-specific manner. Zhang et al.16 studied whole-genome methylation and gene-expression patterns in 153 adult cerebellum samples as a function of the existence of inherited DNA sequence variants. They identified a number of highly significant associations between apparently _cis_- and _trans_-acting SNPs and specific methylation patterns. Many of the SNPs that influenced methylation, and so exhibited allele-specific methylation (ASM), also influenced the expression levels of particular genes. ASM may also influence disease susceptibility, as suggested by Steffanson and colleagues in a study of genetic variants associated with type 2 diabetes17. Other studies suggest that ASE or ASM may be widespread even across different cells within an individual18, 19, although the degree to which this heterogeneity can be attributed to the effect of heterozygous _cis_-acting variants is an open question.
Studies showing widespread ASE and ASM make it clear that the specific DNA sequence and/or epigenetic context associated with each of the two homologous copies of a gene or regulatory element influences the function of these elements in their combined, diploid state. Importantly for the focus of this Opinion article, the effect of ASE and ASM on gene function is likely to be compounded if there are other forms of variation in the same gene (Fig. 1A). A case in point is that of chromogranin A (CHGA), in which common variation in the promoter region has been shown to affect expression and result in ASE. In addition, coding variants have been identified that alter cholinergic inhibition owing to encoded structural deformations that they induce in proteins20. Simply cataloguing the genotypes by combining sequence information from the two chromosomes and ignoring whether heterozygous variants are in cis or trans with other variants would provide incomplete knowledge of an individual’s phenotype with respect to both gene expression and protein function. Thus, the haplotype combinations (diplotype) that an individual possesses are paramount to understanding whether an inhibitory allele is overexpressed or underexpressed relative to the normal allele. Such phenomena are discussed further below in the context of complex disease.
Duplications, deletions and chromosome inequivalence
There is a growing literature on the existence and effect of different numbers of copies of entire genes or parts of genes in individual genomes21, 22. Knowledge of the number of functioning copies of a gene in a single human genome is crucial for determining the potential phenotypic effect of such copy-number variations (CNVs). However, it might be just as important to know how those gene copies are distributed across the two sets of chromosomes in each cell. For example, heterozygous _cis_-acting sequence variations may exist in the surrounding regulatory regions of these gene copies and so influence their function. Thus, the specific combination of gene copies and _cis_-regulatory variants on each chromosomal homologue may dictate the function of those gene copies (Fig. 1B). In this context, it is known that many cancers have somatically acquired ‘amplifications’ in the form of increased copies of particular genes23. Many of these genes have also been found to possess point mutations that influence the function of particular copies23, which may, in turn, influence tumorigenesis24. Understanding the phenotypic effects of deletions also requires knowledge of how variation is partitioned between chromosomes. An example is the phenomenon of ‘unmasking’ potentially deleterious mutations in one copy of a gene when the homologous copy is deleted25 (Fig. 1C).
Diplotypic effects and disease
In addition to the influence of haplotype-specific _cis_-acting variation on gene function in cellular and molecular physiological settings, there have been many documented instances in which specific diplotypes influence disease and clinically relevant phenotypes. We describe examples of such cases below.
Compound heterozygosity
Human disorders often exhibit subtle variation in their phenotypic manifestations. Many studies investigating the genetic mechanisms that underlie this variation, especially in the context of monogenic, overtly Mendelian disorders, have implicated the phenomenon of compound heterozygosity (Table 1). Compound heterozygosity occurs when the two homologous copies of a genomic region each harbour unique sequence variants, but at different positions in that region. These variants are thought to perturb the function of the two homologous copies of a gene in different ways, with their combined molecular effects resulting in a phenotype that is distinct from that seen if one homologous gene carries both deleterious variants26. Thus, in settings in which compound heterozygosity may have a role, merely knowing that an individual is heterozygous for mutations or variants at relevant loci is not enough: knowledge about the specific diplotype is essential.
Table 1.
Example Clinical Conditions and Disorders Influenced by Compound Heterozygosity in Single Genes
| Disease | Reference | Gene Names | Mutations Implicated in Compound Heterozygosity |
|---|---|---|---|
| Blistering Skin | Shimizu59 | COL7A1 | G2316R & G2287R |
| Cerebral Palsy | Fong60 | PROC | N2I & S181R |
| CMT | Lupski9 & McLaughlin61 | SH3TC2 & KARS | SH3TC2: Y169H & R954X, KARS: L133H & Y173SfsX7 |
| Deafness | Welch62 | GJB2 | Additive effect of multiple reported recessive and dominant mutations. |
| Hemachromatosis | Martinez63 | HFE | H63D & 2282Y |
| Mediterranean Fever | Nakamusa64 | MEFV | E14Q & M694I. M694I alone is associated with a mild phenotype |
| Miller Syndrome | Roach4 | DHODH | G152R & G202A |
| Paraganglioma | Majumdar65 | SDHB | V110F & splice donor c.200+7 A > G |
| Hyperphenylalaninemia | Avigad66 | PAH | Multiple PAH variants explained non-PKU HPA cases when acquired as compound heterozygote. |
| FBPase Deficiency | Moon67 | FBP1 | G164S & 838delT |
| Ataxia-telangiectasia | Dörk68 | ATM | Attenuated phenotype: D2625E, A2626P and splice site c.496+5 G>A |
| Glycogen-storage type II | Maimaiti69 | GAA | R600C & splice site c.546G>T. Splice variant has reduced expression |
| Chondrodysplasias | Miyake70 | DTDST | T266I & 340delV |
| Turcot’s Syndrome | De Rosa71 | PMS2 | 1221delG & 2361delCTTC |
Additional instances of clinically relevant compound heterozygosity have been uncovered in large-scale human sequencing studies. For example, Roach et al.4 sequenced the genomes of a pair of siblings with two apparently recessive disorders, Miller syndrome and primary ciliarydyskenesia, and also sequenced the genomes of their parents. Sequence information from the siblings was phased by tracking the transmission of variants from parents to offspring, although not all variants could be unequivocally determined as maternal or paternal in origin. For Miller syndrome, two variants at different positions in the same gene, one on the maternally inherited homologue of the gene and one on the paternally inherited homologue, were proposed to influence the disease. Other instances of compound heterozygosity occur in the context of the ‘two hit’ model of cancer, in which an individual inherits a disruptive cancer-susceptibility variant in one homologue of the gene and then develops a disruptive somatic mutation at a different position in the other homologue. This leads to dysfunction in both gene copies and a potential tumorigenic effect26. It is unclear how often the phenomenon of compound heterozygosity is likely to affect different diseases. However, the fact that there are many known instances in which it does so suggests that studies that use sequencing to identify variants that influence a disease need to take this possibility into account, a task that clearly requires phase information.
Complex diplomic phenomena in common disease
Documented instances of compound heterozygosity have typically involved low-frequency, highly penetrant alleles. It is unclear how such effects relate to the higher-frequency alleles of low effect size that have been shown to contribute considerably to many complex, common disorders over the past few years27. Despite this, some researchers have begun to consider the influence of haplotypic effects in the context of genome-wide association studies investigating common disorders that may reflect compound heterozygosity28, 29. In addition, there is growing evidence for the involvement of specific diplotypes, involving combinations of multiple _cis_-acting variants — some in regulatory regions and some in coding regions — in giving rise to phenotypic effects that contribute to common diseases. The principles discussed above and illustrated in Fig. 1 are also likely to apply in such settings. Table 2 summarizes a range of recently documented instances and we describe some specific examples below.
Table 2.
Example Studies Assessing the Effect of Combinations of Unique Gene-Specific Haplotype Pairs (i.e., Diplotypes) on a Complex Phenotype
| References | Gene | Phenotype assessed | Genetic Basis |
|---|---|---|---|
| Drysdale72 | ADRB2 | Response to asthma therapy | Complex promoter and coding region haplotypes at the ADRB2 locus alter receptor expression. |
| Horan73 | HG1 | HGH expression | Non-additivity of the effects of 16 HG1 SNPs with individual effects, depending on haplotype context. |
| Barroso74 | FANCD2 | Breast cancer | If at least one copy of a specific FANCD2 haplotype is present, carriers are at 4-fold risk. |
| Chen75 | IL1B | IL1B activity | Individual SNP in the IL1B promoter have either an up- or down- regulatory effect depending on haplotype context. |
| Weyrich76 | PRKAG3 | LDL cholesterol | Homozygotes for specific alleles in a specific PRKAG3 diplotype exhibited the highest LDL-cholesterol in among all frequent diplotypes. |
| Yang77 | ATM | Non small lung cancer | Based on haplotype and diplotype analyses a specific diplotype at the ATM locus confers risk. |
| Maggini78 | MDR1 | Multiple myeloma | Protective effects were identified in heterozygotes and homozygotes for a specific diplotype at the MDR1 locus. |
| Pickard79 | NPAS3 | Schizophrenia & bipolar | Combinatorial action of haplotype pairs was associated with overall susceptibility. |
| Sun80 | ADIPOQ | Rosiglitazone response | A specific diplotype at the ADIPOQ locus exhibited stronger association with enhanced response than other diplotypes. |
Two groups identified a strong association between systemic lupus erythematosus (SLE) and haplotypes that contain variants in the protein-coding region of the gene tumour necrosis factor α-induced protein 3 (TNFAIP3)30, 31. Two additional haplotype blocks located ~200 kb upstream and downstream of the TNFAIP3 coding region also showed strong independent signals for association with the disease but were not in linkage disequilibrium with the variants in the coding-region haplotype. The findings raised an important question about how these variants modify autoimmune disease susceptibility in different haplotype conformations. Although neither of the studies explicitly investigated how the variants directly interacted when in cis confirmation, they did provide indirect evidence that the specific diplotype is important.
Graham and colleagues also studied another potential SLE gene, interferon regulatory factor 5 (IRF5)32, 33, 34, which also harbours multiple coding and non-coding variants that exhibit associations with autoimmune diseases. Three separate variants were identified within the IRF5 coding region that disrupt IRF5 function through different mechanisms: abnormal splicing of exon 1b, a 10-residue deletion in exon 6, and disruption of a cleavage and polyadenylation specificity factor (CPSF) site33. Again, an important question is how the distribution of these variants across the two homologous copies of IRF5 in an individual affects overall IRF5 function. For example, the combination of a variant in a splice site and a CPSF mutation on the same chromosome may have a more attenuated effect than if the two variants are on different chromosomes, because in the former case the existence of one functional gene copy with neither variant may compensate for the affected copy with two mutations. Interestingly, Graham and colleagues, and others, have identified further associations implicating additional _cis_-acting regulatory variants in SLE susceptibility33, 34, 35.
A recent example of a complex setting implicating _cis_-acting variants along with structural or repetitive sequences on single chromosomes involved the study of mutations that cause facioscapulohumeral muscular dystrophy36. Here, the contraction of microsatellite repeats has a phenotypic effect only when variants that modify the stability of the double homeobox4 (DUX4) transcript are on the same chromosome as the repeats.
Importance of phase in other settings
In addition to the importance of phase information in resolving how combinations of variants uniquely situated on each homologous genomic region may affect diploid gene function, there are other settings in which phase information is important37. For example, in the context of human population genomic studies, Nievergelt et al. demonstrated that greater differentiation of human populations can be obtained by exploring within- and across-population haplotype diversity than by focusing on multilocus genotype diversity38. In terms of cataloguing human genetic variation, Shendure and colleagues have shown that resolving the existence of structural variants within genomes can be enhanced greatly if phase information is considered37. Studies of the evolution of genomes across species can be enhanced by comparing individual chromosomes39. Finally, classical transplantation studies often exploit haplotype matching to determine optimal host–donor relationships40.
Approaches for diplotyping
Given the importance of knowing the unique nucleotide content associated with each of the two homologous copies of a genomic region for assessing diploid gene function, it is important to consider how this knowledge can be obtained for any individual or group of individuals. There are several approaches for determining phase from DNA sequence and genotype data (Fig. 2). These approaches can be broadly classified in two categories. First, there are methods that leverage genotype information from individuals of either the same population or the same family as a ‘target’ individual whose genome is to be phased. Second, there are methods that physically separate the nucleotide content and unique variants on each homologous chromosome. Importantly, although laboratory and computational methods have the potential to phase or separate two homologous chromosomes, only methods that leverage genotype data from parental lineages can determine whether a particular phased chromosomal copy was inherited from an individual’s mother or father. Knowledge of the specific parental origins of chromosome regions, rather than just the nucleotide content of chromosome homologues, may be of use in the context of parent-of-origin effects such as epigenetic imprinting, as recently demonstrated for type 2 diabetes17.
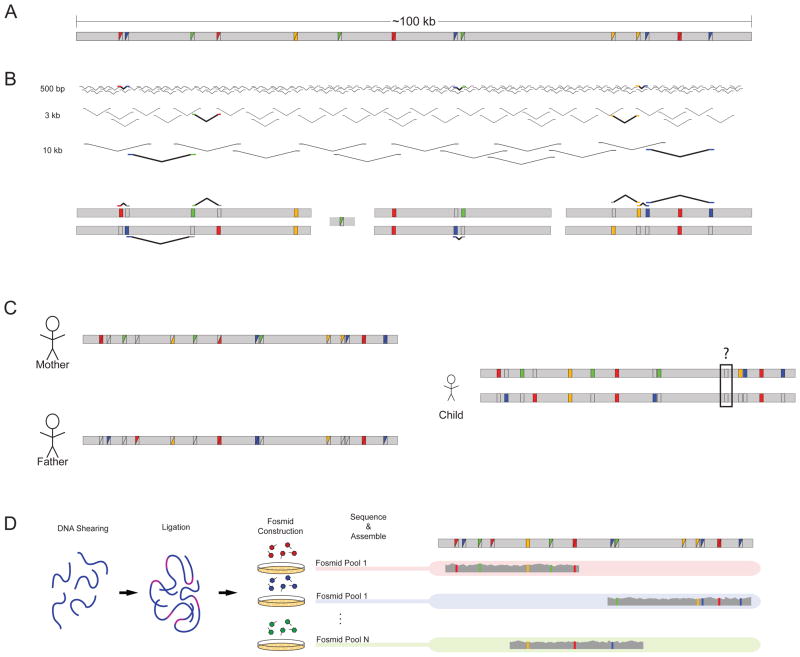
Figure 2. Strategies for empirical haplotype reconstruction.
a | A hypothetical 100 kb stretch of sequence harbours multiple variants compared with the human reference, as designated by the coloured squares. Variants can be homozygous (solid coloured squares) or heterozygous (split coloured squares). b | Sequence reads from libraries of multiple insert sizes can be leveraged to link heterozygous sites together. Informative reads are highlighted and displayed a second time against the diploid reconstruction. The assembly consists of blocks of sequence with gaps arising when variants fall outside the distance of the insert sizes used for sequencing. c | Parental information allows for the separation of chromosomal variants except in instances in which both parents are heterozygous, as demonstrated by the black box in the child’s assembly. d | Laboratory-based methods such as the sequencing of fosmid pools allow for the separation of homologous chromosomes. DNA is sheared, ligated with fosmid vector sequence, packaged and transfected into the bacterium Escherichia coli. Pools of fosmid sequence — each containing only a small fraction of the total genome broken into ~40 kb segments — are sequenced independently. The sequenced libraries are then mapped and assembled for phase reconstruction.
Methods that use information from other individuals
Using information from parents or other relatives is a powerful approach to phasing an individual and has been used in many, if not most, classical family-based human genetic-mapping studies used to identify genomic regions harbouring disease-predisposing variants. Pedigree-based mapping methods such as those that calculate the logarithm of odds (LOD) or that use the transmission disequilibrium test (TDT) track, for example, the transmission of a putative disease-causing variant and a genetic marker together on a single chromosome from generation to generation. Thus, these strategies heavily depend on phase information in the genomic regions of interest. The same approach has been applied to dense genotype data generated by SNP arrays41, as well as whole-genome sequencing (Fig. 2c); for example, in the study by Roach et al.4, discussed above, in which the genomes of two siblings with different Mendelian disorders were sequenced4. Roach et al. reported that by sequencing the parents of the two target individuals, they could separate as much as 96.8% of the genome into maternally and paternally inherited chromosomal or haplotypic complements. Leveraging parental information to phase genomes provides excellent accuracy and demonstrates the added benefit that current family-based genome-sequencing studies will be able to exploit. However, for population- or case–control-based studies this strategy would entail a substantial increase in costs associated with the need to sequence the additional genomes of relatives in addition to those of the target individuals.
The use of genotype data from a larger set of unrelated individuals to phase a target individual can provide a cost-effective method for separating homologous chromosomes with respect to common variants. This approach is based on shared ancestry of the target individual and the larger set of individuals so that linkage-disequilibrium patterns between variants can be exploited in haplotyping the target individual42, 43. However, this approach assumes the availability of genotypes from additional individuals of the same or a similar population as the target and, although the definition of ‘similar’ is often vague, genotype data from individuals of an appropriate population might not be available.
Population-based approaches also assume that there are reliable statistical and computational techniques available to conduct the phasing. Most population-based phasing methods (and related genotype-imputation methods44) can produce reliable haplotypes for moderately long stretches of a chromosome. Human genetics research has a long history of efforts to refine probabilistic phasing methods that leverage data on relatives, entire pedigrees or population linkage-disequilibrium data45, 46 (Table 3). However, these methods are notorious for ‘switching error’ inaccuracies, which arise when chromosomal segments have been phased accurately, but their connections to each other to form larger haplotypes or contigs are incorrect47. Deeper catalogues of genetic variation across many populations may reduce switching errors, but they might be hard to eliminate entirely owing to variation in recombination rates and the genetic diversity within and across human populations. Another problem with the population approach is that it requires the larger set of individuals to have been genotyped previously. As a consequence, these individuals may not be useful for phasing rare variants possessed by the target individual, because rare variants are not likely to have been observed (or may not even exist), and so genotyped, among the larger set of individuals. Hence, reliable linkage-disequilibrium information about those variants might not be available to facilitate phasing. Finally, the population-based phasing approach obviously could not work for private variants possessed only by the target individual.This caveat may be of increasing importance in future studies, as shifts in emphasis begin to focus on understanding rare and even de novo variation and its role in human diseases. In this context, private variants, or variants private to a specific population not previously studied, are unlikely to be accurately phased using data sets such as those associated with the 1000 Genomes Project, given their focus on specific populations48.
Table 3.
Example Methods and Software for Haplotyping and Phasing.
| Method Name | Data Type | Comments |
|---|---|---|
| Hapi81 | Pedigree genotype | Dynamic programming-based haplotype assembly |
| ZRBA82 | Pedigree genotype | Zero-recombination block partition algorithm |
| He et al.58 | Sequencing reads | Dynamic programming-based haplotype assembly |
| HapCut56, 57 | Sequencing reads | Max-Cut based algorithm applicable to arbitrary length reads and insert sizes |
| HASH57 | Sequencing reads | Monte Carlo Markov Chain algorithm for haplotype assembly |
| SHAPE-IT83 | Genotype | Tree representation of hidden markov model |
| Beagle84 | Genotype | Fast and accurate algorithm for phasing using a haplotype-cluster model |
| HaploRec85 | Genotype | Utilizes frequencies of haplotype fragments for phasing |
| fastPHASE86 | Genotype | Haplotype-clustering model for phasing large datasets |
| HAP87 | Genotype | Imperfect phylogeny approach |
| PL-EM88 | Genotype | EM algorithm combined with partition-ligation |
| Merlin89 | Pedigree genotype | Uses sparse gene flow trees to reduce computing requirements |
| Phase90 | Genotype | Most accurate but slow on large datasets |
| Allegro91 | Pedigree genotype | Utilizes multiterminal binary decision diagrams (MTBDDs) for large pedigrees |
| Arlequin92 | Genotype | Expectation-Maximization (EM) algorithm for few markers |
| CRIMAP93 | Pedigree genotype | One of the first pedigree haplotyping programs |
Methods based on information from a single individual
The second set of phasing methods works by seeking to resolve the haplotypic arrangement of two or more neighbouring variants empirically from sequence data gathered on a single individual. Such methods provide a direct approach to phasing and can be used to phase de novo mutations, which, when combined with knowledge of the parental origins of variants surrounding a de novo mutation, can be used to assess, for example, parent-of-origin and paternal age mutation rates, something that is not feasible using other approaches49, 50. Phasing techniques that physically separate chromosomes fall into two broad categories51: separation of complete chromosomes before sequencing, and reduction of the complexity of mixtures of paternally and maternally inherited DNA. Physical separation of entire chromosomes is not trivial because it involves the isolation of chromosomes from a single cell, amplification of the DNA from those isolated chromosomes, and then sequencing. The use of sophisticated microfluidic technologies has recently been applied to this process40 and represents a substantial improvement over previous methods52.
Complexity reduction involves the separation of genomic DNA into pools that contain DNA from regions of the genome that are either maternally or paternally derived53. A compelling recent example of this approach used 115 fosmid libraries to reconstruct the diploid sequence of the genome of a South Asian individual37 (Fig. 2d). As an alternative to the use of fosmid libraries, pooled maternal and paternal DNA samples diluted to a point at which only a fraction of a complete genome is present for sequencing could be used. With the proper assessment of the dilutions, each pool will be expected to contain only a single chromosome at any particular region54. Cloning- and dilution-based methods for complexity reduction are straightforward and probably within the capabilities of most sequencing laboratories with standard equipment, but result only in large contigs that reflect haplotypic segments of a chromosome that still need to be stitched together to characterize an entire chromosome — a process that could be error prone.
As an alternative, phase can be reconstructed from diploid DNA from a single individual using computational approaches that link partially overlapping DNA-sequencing reads harbouring variants at heterozygous positions55, 56, 57, 58 (Fig. 2b). This approach requires long DNA-sequencing reads or mate-pairs of variable insert size in order to reliably capture multiple heterozygous sites that can be used to assemble reads into larger contigs on the basis of their overlapping nucleotide content56. This approach was used in the construction of the first diploid genome1, although, owing to limitations in the available sequence data and the number of heterozygous positions spanned by the sequencing reads, only ~70% of the genome could be phased. Current sequencing projects that use a limited selection of short insert size, paired-read distances are not well designed for phase reconstruction. Future work should focus on improvements to mate-pair construction and projects that leverage variable insert size libraries, which, coupled with longer reads, should allow reasonably sized haploid contig assemblies (Fig. 3).
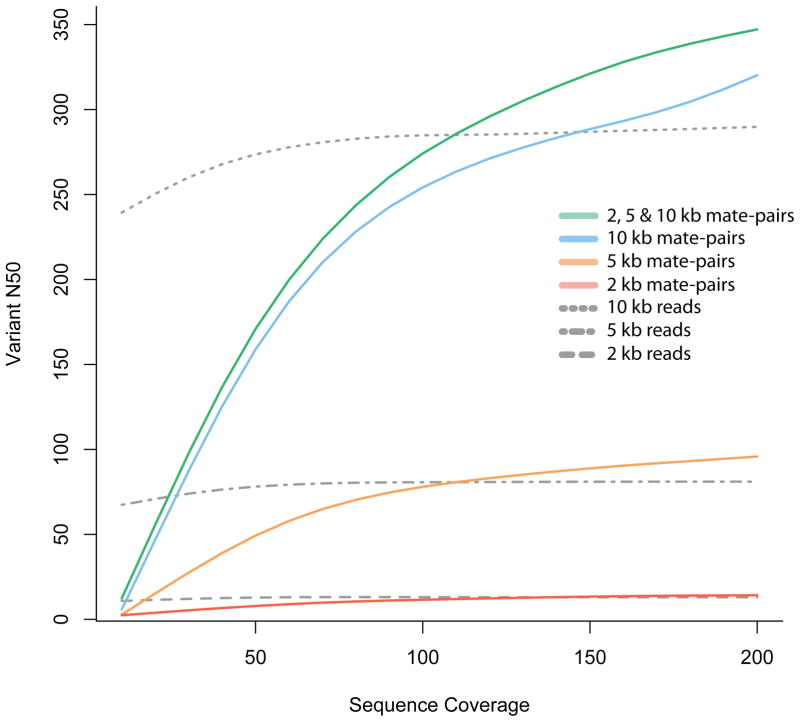
Figure 3. Phase reconstruction using mate-pair information.
Simulated 100 bp mate-pair read coverage of various depths (sequence (fold) coverage, _x_-axis) for chromosome 1 of a Yoruban individual. All simulations were done using SNP calls (for chromosome 1) for the Yoruban individual NA19240, obtained from the 1000 Genomes project (released December 2008). Paired-end reads were simulated with the starting position of one read, chosen consistently at random on the chromosome, and the insert length sampled from a normal distribution with a given mean insert length (2, 5 or 10 kb) and standard deviation equal to 10% of the mean. For each simulation experiment, we constructed a graph with nodes corresponding to the heterozygous SNPs and edges corresponding to reads that cover multiple variants. The N50 was calculated using the number of variants in each connected component of this graph that correspond to the phased haplotype blocks. The vN50 is defined as the point at which half of the heterozygous loci of the chromosome are contained in contigs with the vN50 or greater number of variants. Mate-pair libraries outperform reads of the same length because the size distribution of the insert consists of lengths greater than 10 kb, allowing for longer connections than are possible with single reads alone.
Diplomics: a new frontier?
We have emphasized why an understanding of how specific combinations of genetic variants on the two homologous copies of a chromosomal region influence diploid gene function is crucial for human genetic research. There may, however, be other phenomena that reflect the consequences of diploidy that we have not touched on here. For example, differences in the mere lengths of inherited genomes (owing to, for example, copy-number variations, repeat polymorphisms or large indels) may affect DNA packing and epigenomic phenomena. For these reasons, the science of diplomics should receive greater attention in the human genetics community in the future. However, as we have argued, diplomic enquiry requires more sophisticated sequencing and study-design strategies than those in current use. For example, better a priori chromosome-separation techniques are needed for human sequencing studies, as are sequencing technologies that generate longer reads to facilitate de novo haplotype-based assemblies. We foresee that a re-emergence of family studies will occur to help to resolve important diplomics-related issues, such as those involving complex forms of compound heterozygosity. Finally, in order to fully understand how the diplotypic genomic ‘whole’ functions over and above its haplotypic ‘parts’, we believe that more relevant functional assays, perhaps involving the simultaneous introduction of different haplotypic complements into functional assays or transgenic animals, are needed. Ultimately, if collaborative science teams are to make headway in unravelling the secrets of the human genome, especially in refining the functional and clinical effects of human genomic variation, then it makes no sense to ignore one of its most fundamental aspects: its diploid nature.
Acknowledgments
This work was supported, in part, by the following research grants: U19 AG023122-01, R01 MH078151-01A1, N01 MH22005, U01 DA024417-01, P50 MH081755-01 and UL1 RR025774, as well as the Price Foundation and Scripps Genomic Medicine. This work is the authors’ sole responsibility and does not necessarily represent funding agencies’ views.
References
- 1.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lifton RP. Individual genomes on the horizon. N Engl J Med. 2010;362:1235–1236. doi: 10.1056/NEJMe1001090. [DOI] [PubMed] [Google Scholar]
- 3.Ashley EA, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nature Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lupski JR, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montgomery SB, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morozova O, Hirst M, Marra MA. Applications of new sequencing technologies for transcriptome analysis. Annu Rev Genomics Hum Genet. 2009;10:135–151. doi: 10.1146/annurev-genom-082908-145957. [DOI] [PubMed] [Google Scholar]
- 13.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nature Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tucker T, Marra M, Friedman JM. Massively parallel sequencing: the next big thing in genetic medicine. Am J Hum Genet. 2009;85:142–154. doi: 10.1016/j.ajhg.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDaniell R, et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D, et al. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411–419. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong A, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tycko B. Mapping allele-specific DNA methylation: a new tool for maximizing information from GWAS. Am J Hum Genet. 2010;86:109–112. doi: 10.1016/j.ajhg.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 20.Wen G, et al. Both rare and common polymorphisms contribute functional variation at CHGA, a regulator of catecholamine physiology. Am J Hum Genet. 2004;74:197–207. doi: 10.1086/381399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nature Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wain LV, Armour JA, Tobin MD. Genomic copy number variation, human health, and disease. Lancet. 2009;374:340–350. doi: 10.1016/S0140-6736(09)60249-X. [DOI] [PubMed] [Google Scholar]
- 23.Leary RJ, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci USA. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudson AG. Two genetic hits (more or less) to cancer. Nature Rev Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 25.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nature Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 26.Zschocke J. Dominant versus recessive: molecular mechanisms in metabolic disease. J Inherit Metab Dis. 2008;31:599–618. doi: 10.1007/s10545-008-1016-5. [DOI] [PubMed] [Google Scholar]
- 27.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nature Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 28.Su Z, Cardin N, Donnelly P, Marchini J, Control WTC. A Bayesian method for detecting and characterizing allelic heterogeneity and boosting signals in genome-wide association etudies. Statistical Sci. 2009;24:430–450. [Google Scholar]
- 29.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. Plos Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham RR, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nature Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musone SL, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nature Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham RR, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nature Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 33.Graham RR, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci USA. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harley JB, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nature Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimane K, et al. The association of a nonsynonymous single-nucleotide polymorphism in TNFAIP3 with systemic lupus erythematosus and rheumatoid arthritis in the Japanese population. Arthritis Rheum. 2010;62:574–579. doi: 10.1002/art.27190. [DOI] [PubMed] [Google Scholar]
- 36.Lemmers RJ, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitzman JO, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nature Biotech. 2010 Dec 19; doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nievergelt CM, Libiger O, Schork NJ. Generalized analysis of molecular variance. PLoS Genet. 2007;3:e51. doi: 10.1371/journal.pgen.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nature Biotech. 2010 Dec 19; doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong A, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nature Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Browning SR. Missing data imputation and haplotype phase inference for genome-wide association studies. Hum Genet. 2008;124:439–450. doi: 10.1007/s00439-008-0568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biernacka JM, et al. Assessment of genotype imputation methods. BMC Proc. 2009;3(Suppl 7):S5. doi: 10.1186/1753-6561-3-s7-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao G, Allison DB, Hoeschele I. Haplotyping methods for pedigrees. Hum Hered. 2009;67:248–266. doi: 10.1159/000194978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salem RM, Wessel J, Schork NJ. A comprehensive literature review of haplotyping software and methods for use with unrelated individuals. Hum Genomics. 2005;2:39–66. doi: 10.1186/1479-7364-2-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andres AM, et al. Understanding the accuracy of statistical haplotype inference with sequence data of known phase. Genet Epidemiol. 2007;31:659–671. doi: 10.1002/gepi.20185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goriely A, Wilkie AO. Missing heritability: paternal age effect mutations and selfish spermatogonia. Nature Rev Genet. 2010;11:589. doi: 10.1038/nrg2809-c1. [DOI] [PubMed] [Google Scholar]
- 50.Moloney DM, et al. Exclusive paternal origin of new mutations in Apert syndrome. Nature Genet. 1996;13:48–53. doi: 10.1038/ng0596-48. [DOI] [PubMed] [Google Scholar]
- 51.Bansal V, Tewhey R, Topol EJ, Schork N. The next phase in human genetics. Nature Biotech. 2011;29:38–39. doi: 10.1038/nbt.1757. [DOI] [PubMed] [Google Scholar]
- 52.Ma L, et al. Direct determination of molecular haplotypes by chromosome microdissection. Nature Methods. 2010;7:299–301. doi: 10.1038/nmeth.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kouprina N, Larionov V. TAR cloning: insights into gene function, long-range haplotypes and genome structure and evolution. Nature Rev Genet. 2006;7:805–812. doi: 10.1038/nrg1943. [DOI] [PubMed] [Google Scholar]
- 54.Paul P, Apgar J. Single-molecule dilution and multiple displacement amplification for molecular haplotyping. Biotechniques. 2005;38:553–559. doi: 10.2144/05384ST01. [DOI] [PubMed] [Google Scholar]
- 55.Kim JH, Waterman MS, Li LM. Diploid genome reconstruction of Cionaintestinalis and comparative analysis with. Cionasavignyi Genome Res. 2007;17:1101–1110. doi: 10.1101/gr.5894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bansal V, Bafna V. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics. 2008;24:i153–159. doi: 10.1093/bioinformatics/btn298. [DOI] [PubMed] [Google Scholar]
- 57.Bansal V, Halpern AL, Axelrod N, Bafna V. An MCMC algorithm for haplotype assembly from whole-genome sequence data. Genome Res. 2008;18:1336–1346. doi: 10.1101/gr.077065.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He D, Choi A, Pipatsrisawat K, Darwiche A, Eskin E. Optimal algorithms for haplotype assembly from whole-genome sequence data. Bioinformatics. 2010;26:i183–i190. doi: 10.1093/bioinformatics/btq215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimizu H, et al. Epidermolysis bullosa simplex associated with muscular dystrophy: phenotype-genotype correlations and review of the literature. J Am Acad Dermatol. 1999;41:950–956. doi: 10.1016/s0190-9622(99)70252-5. [DOI] [PubMed] [Google Scholar]
- 60.Fong CY, Mumford AD, Likeman MJ, Jardine PE. Cerebral palsy in siblings caused by compound heterozygous mutations in the gene encoding protein C. Dev Med Child Neurol. 2010;52:489–493. doi: 10.1111/j.1469-8749.2010.03618.x. [DOI] [PubMed] [Google Scholar]
- 61.McLaughlin HM, et al. Compound heterozygosity for loss-of-function lysyl-tRNAsynthetase mutations in a patient with peripheral neuropathy. Am J Hum Genet. 2010;87:560–566. doi: 10.1016/j.ajhg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Welch KO, Marin RS, Pandya A, Arnos KS. Compound heterozygosity for dominant and recessive GJB2 mutations: effect on phenotype and review of the literature. Am J Med Genet A. 2007;143A:1567–1573. doi: 10.1002/ajmg.a.31701. [DOI] [PubMed] [Google Scholar]
- 63.Aguilar Martinez P, et al. Compound heterozygotes for hemochromatosis gene mutations: may they help to understand the pathophysiology of the disease? Blood Cells Mol Dis. 1997;23:269–276. doi: 10.1006/bcmd.1997.0143. [DOI] [PubMed] [Google Scholar]
- 64.Nakamura A, Yazaki M, Tokuda T, Hattori T, Ikeda S. A Japanese patient with familial Mediterranean fever associated with compound heterozygosity for pyrin variant E148Q/M694I. Intern Med. 2005;44:261–265. doi: 10.2169/internalmedicine.44.261. [DOI] [PubMed] [Google Scholar]
- 65.Majumdar S, et al. Compound heterozygous mutation with a novel splice donor region DNA sequence variant in the succinate dehydrogenase subunit B gene in malignant paraganglioma. Pediatr Blood Cancer. 2010;54:473–475. doi: 10.1002/pbc.22338. [DOI] [PubMed] [Google Scholar]
- 66.Avigad S, et al. Compound heterozygosity in nonphenylketonuria hyperphenylalanemia: the contribution of mutations for classical phenylketonuria. Am J Hum Genet. 1991;49:393–399. [PMC free article] [PubMed] [Google Scholar]
- 67.Moon S, et al. Novel compound heterozygous mutations in the fructose-1,6-bisphosphatase gene cause hypoglycemia and lactic acidosis. Metabolism. 2011;60:107–113. doi: 10.1016/j.metabol.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 68.Dork T, Bendix-Waltes R, Wegner RD, Stumm M. Slow progression of ataxia-telangiectasia with double missense and in frame splice mutations. Am J Med Genet A. 2004;126A:272–277. doi: 10.1002/ajmg.a.20601. [DOI] [PubMed] [Google Scholar]
- 69.Maimaiti M, et al. Silent exonic mutation in the acid-α-glycosidase gene that causes glycogen storage disease type II by affecting mRNA splicing. J Hum Genet. 2009;54:493–496. doi: 10.1038/jhg.2009.66. [DOI] [PubMed] [Google Scholar]
- 70.Miyake A, et al. A compound heterozygote of novel and recurrent DTDST mutations results in a novel intermediate phenotype of Desbuquois dysplasia, diastrophic dysplasia, and recessive form of multiple epiphyseal dysplasia. J Hum Genet. 2008;53:764–768. doi: 10.1007/s10038-008-0305-z. [DOI] [PubMed] [Google Scholar]
- 71.De Rosa M, et al. Evidence for a recessive inheritance of Turcot’s syndrome caused by compound heterozygous mutations within the PMS2 gene. Oncogene. 2000;19:1719–1723. doi: 10.1038/sj.onc.1203447. [DOI] [PubMed] [Google Scholar]
- 72.Drysdale CM, et al. Complex promoter and coding region β2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci USA. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horan M, et al. Human growth hormone 1 (GH1) gene expression: complex haplotype-dependent influence of polymorphic variation in the proximal promoter and locus control region. Hum Mutat. 2003;21:408–423. doi: 10.1002/humu.10167. [DOI] [PubMed] [Google Scholar]
- 74.Barroso E, et al. FANCD2 associated with sporadic breast cancer risk. Carcinogenesis. 2006;27:1930–1937. doi: 10.1093/carcin/bgl062. [DOI] [PubMed] [Google Scholar]
- 75.Chen H, et al. Single nucleotide polymorphisms in the human interleukin-1B gene affect transcription according to haplotype context. Hum Mol Genet. 2006;15:519–529. doi: 10.1093/hmg/ddi469. [DOI] [PubMed] [Google Scholar]
- 76.Weyrich P, et al. Role of AMP-activated protein kinase gamma 3 genetic variability in glucose and lipid metabolism in non-diabetic whites. Diabetologia. 2007;50:2097–2106. doi: 10.1007/s00125-007-0788-8. [DOI] [PubMed] [Google Scholar]
- 77.Yang H, et al. ATM sequence variants associate with susceptibility to non-small cell lung cancer. Int J Cancer. 2007;121:2254–2259. doi: 10.1002/ijc.22918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maggini V, et al. MDR1 diplotypes as prognostic markers in multiple myeloma. Pharmacogenet Genomics. 2008;18:383–389. doi: 10.1097/FPC.0b013e3282f82297. [DOI] [PubMed] [Google Scholar]
- 79.Pickard BS, et al. Interacting haplotypes at the NPAS3 locus alter risk of schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14:874–884. doi: 10.1038/mp.2008.24. [DOI] [PubMed] [Google Scholar]
- 80.Sun H, et al. The association of adiponectin allele 45T/G and -11377C/G polymorphisms with type 2 diabetes and rosiglitazone response in Chinese patients. Br J Clin Pharmacol. 2008;65:917–926. doi: 10.1111/j.1365-2125.2008.03145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams AL, Housman DE, Rinard MC, Gifford DK. Rapid haplotype inference for nuclear families. Genome Biol. 2010;11:R108. doi: 10.1186/gb-2010-11-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang HT, Xu Y, Zhao YZ, Chen GL. A novel algorithm for minimum recombinant haplotyping on pedigrees by zero recombinant block partition. Interdiscip Sci. 2010;2:185–192. doi: 10.1007/s12539-010-0089-7. [DOI] [PubMed] [Google Scholar]
- 83.Delaneau O, Coulonges C, Zagury JF. Shape-IT: new rapid and accurate algorithm for haplotype inference. BMC Bioinformatics. 2008;9:540. doi: 10.1186/1471-2105-9-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eronen L, Geerts F, Toivonen H. HaploRec: efficient and accurate large-scale reconstruction of haplotypes. BMC Bioinformatics. 2006;7:542. doi: 10.1186/1471-2105-7-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Halperin E, Eskin E. Haplotype reconstruction from genotype data using imperfect phylogeny. Bioinformatics. 2004;20:1842–1849. doi: 10.1093/bioinformatics/bth149. [DOI] [PubMed] [Google Scholar]
- 88.Qin ZS, Niu T, Liu JS. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet. 2002;71:1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nature Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 90.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gudbjartsson DF, Thorvaldsson T, Kong A, Gunnarsson G, Ingolfsdottir A. Allegro version 2. Nature Genet. 2005;37:1015–1016. doi: 10.1038/ng1005-1015. [DOI] [PubMed] [Google Scholar]
- 92.Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 93.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]