Infantile-Onset Ascending Hereditary Spastic Paralysis Is Associated with Mutations in the Alsin Gene (original) (raw)
Abstract
We studied 15 patients, from 10 families, who presented with severe spastic paralysis with an infantile onset and an ascending progression. Spastic paraplegia began during the first 2 years of life and extended to upper limbs within the next few years. During the first decade of life, the disease progressed to tetraplegia, anarthria, dysphagia, and slow eye movements. Overall, the disease was compatible with long survival. Signs of lower motor-neuron involvement were never observed, whereas motor-evoked potentials and magnetic resonance imaging demonstrated a primitive, pure degeneration of the upper motor neurons. Genotyping and linkage analyses demonstrated that this infantile-onset ascending hereditary spastic paralysis (IAHSP) is allelic to the condition previously reported as juvenile amyotrophic lateral sclerosis at the ALS2 locus on chromosome 2q33-35 (LOD score 6.66 at recombination fraction 0). We analyzed ALS2, recently found mutated in consanguineous Arabic families presenting either an ALS2 phenotype or juvenile-onset primary lateral sclerosis (JPLS), as a candidate gene. In 4 of the 10 families, we found abnormalities: three deletions and one splice-site mutation. All the mutations lead to a truncated alsin protein. In one case, the mutation affected both the short and the long alsin transcript. In the six remaining families, absence of cDNA ALS2 mutations suggests either mutations in regulatory ALS2 regions or genetic heterogeneity, as already reported in JPLS. Alsin mutations are responsible for a primitive, retrograde degeneration of the upper motor neurons of the pyramidal tracts, leading to a clinical continuum from infantile (IAHSP) to juvenile forms with (ALS2) or without (JPLS) lower motor-neuron involvement. Further analyses will determine whether other hereditary disorders with primitive involvement of the central motor pathways, as pure forms of spastic paraplegia, could be due to alsin dysfunction.
Introduction
Hereditary spastic paraplegia (HSP) and amyotrophic lateral sclerosis (ALS) are two groups of disorders characterized mainly by primary degeneration of the central motor pathways. HSP encompasses a heterogeneous and expanding group of conditions whose prominent clinical features are progressive spasticity and weakness of lower limbs, which may or may not be associated with additional symptoms (Harding 1981). The genetic heterogeneity of pure HSP is known, with an increasing number of chromosomal localizations (Figlewicz and Bird 1999; Seri et al. 1999; McDermott et al. 2000; Reid et al. 2000; Vazza et al. 2000). Six genes have been identified: spastin (MIM 604277; or SPG4, MIM 182601) (Hazan et al. 1999), paraplegin (MIM 604277; or SPG7, MIM 602783) (Casari et al. 1998), L1CAM (MIM 308840; or SPG1, MIM 312900) (Jouet 1994), PLP (MIM 312080; or SPG2, MIM 312920) (Saugier-Veber 1994), atlastin (MIM 606439; or SPG3A, MIM 182600) (Zhao et al. 2001), and HSP60 (MIM 118190; or SPG13, MIM 118190) (Hansen 2002). Amyotrophic lateral sclerosis (ALS) is a distinct condition, involving both upper and lower motor neurons, that results in a progressive ascending paralysis of lower and upper limbs and cranial nerves. The classical adult form is sporadic and rapidly leads to death 2–5 years after onset (Mulder 1986). Rare familial forms with a dominant or recessive pattern of transmission have been described. Autosomal dominant forms have been mapped to chromosome 21q21 (ALS1 [MIM 105400]) (Siddique 1991) and 18q21 (ALS6 [MIM 606640]) (Hand et al. 2002). Patients with ALS1 harbor mutations in the superoxide dismutase gene (SOD1 [MIM 147450] (Rosen et al. 1993). Families with more rare autosomal recessive inheritance have shown linkage to several loci: 2q33 (ALS2 [MIM 205100]), 15q15-21 (ALS5 [MIM 602099]), and 9q34 (ALS4 [MIM 602433]) (Hentati et al. 1994, 1998; Chance et al. 1998]. Patients from a large consanguineous Tunisian family with juvenile-onset ALS show linkage to the 2q33-35 locus (ALS2) (Hentati et al. 1994, 1998; Chance et al. 1998).
The term “primary lateral sclerosis” (PLS) has been used to describe a very uncommon type of selective upper motor-neuron involvement, sparing lower motor neurons. Its relationship with ALS remains controversial (Pringle et al. 1992). Recently, homozygous mutations in a newly identified gene (ALS2 [MIM 606352]) on chromosome 2q33 have been found in the initial Tunisian family with ALS2 and in three additional consanguineous families from the Arabic peninsula. One family displays a juvenile-onset ALS2 phenotype, and two families display a juvenile-onset PLS phenotype (PLSJ [MIM 606353]; Hadano et al. 2001_a_; Yang et al. 2001). The ALS2 gene encompasses 34 exons and encodes a protein termed “alsin.” Two alternative spliced variants have been described: the long variant (6394 nt, 1657 aa) and the short variant (2651 nt, 396 aa), the latter resulting in a premature stop codon after 25 aa residues in intron 4. It has been proposed that ALS2 mutations might predict the severe PLS phenotype, when only the long alsin protein is altered, or the even more severe ALS phenotype, when the short transcript is also affected. Herein we report genetic analyses of 15 patients from 10 unrelated families who presented a pure form of slow, progressive, ascending spastic paralysis with involvement of the lower limbs during infancy (age 1–2 years) and subsequent extension to the upper limbs and bulbar muscles after 5–10 years of evolution.
Subjects and Methods
Patients
We studied 15 patients from 10 unrelated families of different ethnic origins (fig. 1 and table 1). In four families, affected sib pairs suggested a recessive pattern of transmission (families 362, 408, 279, and 786). In family 362, from Algeria, parents were first cousins. In family 408, both parents were members of the same Libyan tribe with a strong history of consanguinity; however, a precise pedigree to identify the degree of consanguinity between both parents was impossible to obtain. In families 279 and 786, of Italian origin, no consanguinity was reported. In six families, there was a single affected child and healthy siblings: one consanguineous family was from Italy (family 283) and five nonconsanguineous families were from France (families 419 and 242) and Italy (families 278, 747, and 786). In all cases, clinical examination of the parents was normal, and no history of neurological disease in the ascendants was reported.
Figure 1.
Pedigree and haplotype analysis of 10 families with the IAHSP phenotype. Individuals from 10 families were genotyped with 15 markers of the 2q33-34 region, including the markers of the ALS2 critical region (D2S116, D2S2309, D2S2214, D2S346, D2S2289, D2S72, D2S307, D2S2189, and D2S2237). Haplotypes were generated under the assumption that the smallest number of recombination events was present. Haplotype analysis defined the boundary of the IAHSP locus within the ALS2 critical region, between markers D2S116 and D2S2237. For ALS2, a plus sign (+) denotes the mutated allele and a minus sign (−) the nonmutated allele. Asterisks (*) denote families with an ALS2 mutation.
Table 1.
Summary of Clinical and MRI Data for 15 Patients with AIHSP
| FamilyandPatient | Countryof Origin | Sex | Age(years) | Age atOnset(years) | Loss ofWalking(years)a | Upper-LimbInvolvment(years) | BulbarInvolvement(years) | AlsinMutationFound |
|---|---|---|---|---|---|---|---|---|
| 362: | Algeria | |||||||
| 2 | M | 36 | 1 | NA | <7 | 13 | Yes | |
| 3 | F | 31 | 1 | NA | <7 | 13 | Yes | |
| 5 | M | 24 | 1 | NA | <7 | 13 | Yes | |
| 279: | Italy | |||||||
| 1 | M | 14 | 1.2 | 1.5 | 9 | 10 | No | |
| 2 | M | 10 | 1 | 10 | 7 | 10 | No | |
| 408: | Libya | |||||||
| 1 | M | 8 | 1.5 | 8 | No | No | No | |
| 2 | M | 16 | 1 | 10 | No | 15 | No | |
| 419-1 | France | F | 18 | 1.5 | 4 | 6 | 8 | Yes |
| 283-1 | Italy | F | 23 | 1.4 | 5 | 10 | 12 | Yes |
| 278-1 | Italy | M | 20 | 1.5 | 4 | 9 | 13 | Yes |
| 242-2 | France | M | 18 | 5 | 9 | 13 | 16 | No |
| 751: | Italy | |||||||
| 1 | M | 11 | 1.5 | NA | 7 | 10 | No | |
| 2 | F | 4 | 2 | No | No | No | No | |
| 747-2 | Italy | F | 20 | 1 | 4 | 10 | 11 | No |
| 786-2 | Italy | F | 7 | 3 | 4 | No | No | No |
All patients presented a severe spastic paralysis with an infantile onset and a stereotyped ascending progression leading to tetraplegia and anarthria during the early teenage years. All were considered normal at birth, and spastic paraplegia started approximately at the age of walking achievement. Weakness and spasticity, with signs of pyramidal tract involvement, extended to the upper limbs at age ∼7–10 years. All the patients were wheelchair-bound by the age of 10 years. In the second decade, the disease progressed to tetraplegia, anarthria, dysphagia, and slow eye movements. Despite this very early onset, long survival beyond the third and even fourth decade was observed.
No other neurological or extraneurological clinical features were noticed, and cognitive functions were spared. Results of extensive studies, including a wide range of metabolic studies, muscular biopsy, and electromyography (EMG) with sensory and motor nerve–conduction velocities (NCV), were normal. Motor-evoked potentials (MEP) in all patients showed early abolition of corticospinal responses, in contrast with normal responses of the somatosensory-evoked potentials (SSEP), which became abnormal only in the later stages. Visual and auditory evoked potentials were also normal. Brain MRI showed different degrees of abnormalities, extending from normal images in the youngest patients to severe sylvian, brainstem, and spinal cord atrophy with hyperlucencies in T2-weighted images along the pyramidal tract in the oldest patients.
Genotyping
Informed consent was obtained from all families, in accordance with local institutional review board guidelines. Genomic DNA from peripheral blood was extracted with Nucleon BACC2 (Amersham). Forty-three polymorphic microsatellites were used for genotyping. Twenty-eight microsatellites were in known HSP loci: 10 for autosomal recessive loci (8q11-12 [D8S285 and D8S260], 16q24.3 [D16S520, D16S413, and D16S3023], 15q13 [D15S1007, D15S971, and D15S1012], and 3q27-28 [D3S1294 and D3S2747]) and 18 for autosomal dominant loci (2p21 [D2S367], 14q13-21 [D14S288 and D14S276], 15pter-q14 [D15S128, D15S1002, and D15S165], 8q23-24 [D8S514 and D8S284], 19q13 [D19S220, D19S420, and D19S902], 12q13 [D12S368, D12S1586, and D12S83], 2q24 [D2S117], and 10q23.3-24.2 [D10S192, D10S1709, and D10S1686]). Fifteen microsatellites were located in and around the ALS2 locus on chromosome 2q33-35: D2S117, D2S115, D2S116, D2S309, D2S2309, D2S2214, D2S346, D2S2289, D2S72, D2S307, D2S2189, D2S2237, D2S155, D2S325, and D2S2382.
PCR amplification and electrophoresis
Primers for polymorphic markers D8S285, D8S260, D16S520, D15S1007, D15S1012, D2S367, D14S288, D14S276, D15S128, D15S1002, D15S165, D8S514, D8S284, D19S220, D19S420, D19S902, D12S368, D12S83, D10S192, D2S117, D2S325, and D2S2382 were obtained from the ABI Linkage Mapping Set version 2 (LMS2; Applied Biosystems) and were used according to the conditions recommended by the manufacturer. Primer sequences of the remaining 15 markers—D2S116, D2S115, D2S2309, D2S2214, D2S309, D2S307, D2S346, D2S2289, D2S72, D2S2189, D2S2237, D2S155, D16S413, D16S3023, D15S971, D3S1294, D3S2747, D12S1586, and D10S1709—were available from the Généthon microsatellite linkage map. PCR reactions and thermocycling conditions were identical to those used for markers from the LMS2 marker set. PCR products were electrophoresed on a 4.25% denaturing polyacrylamide gel, using an ABI 377 sequencer. They were sized by the Genescan program, version 2.1, and scored by the Genotyper 2.0 program.
Linkage analysis
Linkage analyses were performed on a Sun station, using the facilities provided by the INFOBIOGEN support. The LINKAGE package [Lathrop et al. 1985], version 5.1, was used for the generation of pedigree files (MAKEPED) and data files (PREPLINK), using the allele frequency of each marker as found in the CEPH database.
Two-point LOD scores were calculated by MLINK, under the assumption of recessive inheritance with a penetrance of 90%. Recombination frequencies were assumed to be equal between males and females.
Multipoint LOD scores were calculated for 15 markers on chromosome 2q33-35 with GENEHUNTER, version 2.0, assuming a recessive mode of inheritance, genetic homogeneity, and a penetrance of 90%. Allele frequencies of each marker were those of the CEPH database. The order of markers and their respective distances were obtained from linkage maps from the Généthon microsatellite linkage map and the Marshfield Center for Medical Genetics and from the physical map of the ALS2 critical region on human chromosome 2q33-34 (Hadano et al. 1999), as follows: D2S117–1.2 cM–D2S115–3 cM–D2S116–0.1 cM–D2S309–0.1 cM–D2S2309–0.1 cM–D2S2214–0.1 cM–D2S346–0.53 cM–D2S2289–0.1 cM–D2S72–0.53 cM–D2S307–0.1 cM–D2S2189–1.25 cM–D2S2237–2.49 cM–D2S155–1.61 cM–D2S325–8.96 cM–D2S2382.
Mutation Detection
Total RNA was extracted from Epstein-Barr virus (EBV)–transformed lymphoblasts from all affected patients, with RNA Now (Biogentex). First-strand cDNA synthesis with an oligo-dT primer (Superscript Preamplification system [Invitrogen]) was followed by PCR amplification with the 17 primers listed in table 2, which cover the short as well as the long ALS2 cDNA. After 35 cycles of PCR amplification, we purified the PCR products, using a QIAquick PCR purification kit (Qiagen). We sequenced both strands of PCR products, using the Big DyeDeoxy Terminator Cycle Sequencing kit (Perkin Elmer) on an ABI 377 automatic sequencer (Applied Biosystems).
Table 2.
Sequences of Primers for PCR and ALS2 cDNA Sequencing
| Primer (5′→3′) | |||
|---|---|---|---|
| PrimerSetName | Forward | Reverse | ProductSize(bp) |
| ALS2-1 | ATGATTTGGTGATGG | GACAGGATTTGGTTC | 428 |
| ALS2-2 | GGAGCAGTGACAGAC | CCTAATGGGCAACAA | 455 |
| ALS2-3 | CAATGCCTCCCTTCC | ATGGTATGTTTCTGG | 346 |
| ALS2-4 | GCAGGAAGAAGTATT | AAAACAAGCCCAACA | 481 |
| ALS2-5 | GTCCTCTCAACAAAA | TCTGCTTCTCCACTG | 585 |
| ALS2-6 | AGGCAGGCAGTAGTG | TGGGATTTCGCAGTA | 418 |
| ALS2-7 | CTGACCCCACATAC | TCCACCAAGGCTAAA | 515 |
| ALS2-8 | CCCAGGTTTACTCAT | CAATGGCAGGCTTAG | 672 |
| ALS2-9 | ACAACCTTCCAGAGA | GCTTCCTTCCTTTT | 712 |
| ALS2-10 | AAGATTTGTCAGAAG | TGGTGCGTGGAGAA | 391 |
| ALS2-11 | GTCTTGCTCTCCATC | CCATACCCATCTTCC | 610 |
| ALS2-12 | TCACTCTCATTTCAT | TTCTTATCCATTACC | 517 |
| ALS2-13 | TTGGAAGATGGGTAT | TAGGTTTGAAGTAGG | 547 |
| ALS2-14 | GAATGTGGCAAGATG | TCAGGAAATAAGAAC | 751 |
| ALS2-15 | TCACTGGAACCTACT | GCATAAAGCATAAAC | 691 |
| ALS2-16 | CTGGTGAGGTTCTTA | GCATCTTTCGTGGTT | 362 |
| ALS2-17 | GCTGTTTATGCTTTA | GCTCTTATTATTGGT | 649 |
When mutations were detected, the corresponding exons were amplified from genomic DNA of all family members, using the primers and conditions proposed by Hadano et al. (2001_a_). Sequencing conditions were identical to those used in the cDNA analysis.
Results
Linkage and Haplotype Analysis
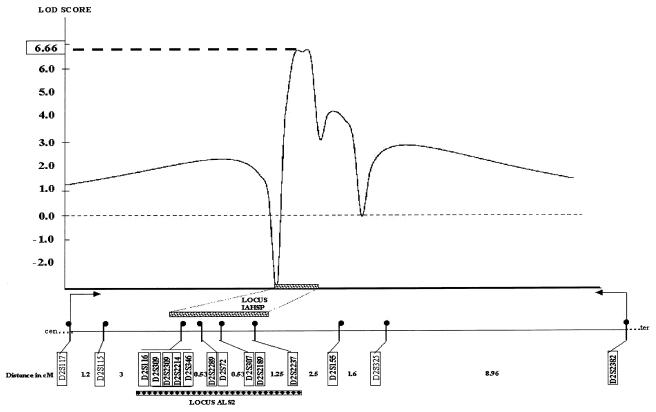
Genotyping for the 12 recessive and dominant HSP loci reported was performed in seven families (families 362, 279, 408, 419, 283, 278, and 242). These loci were excluded by a two-point LOD score ⩽0 for all families and by haplotype analysis, which showed recombinant events (data not shown). Using 11 markers (D2S117, D2S116, D2S309, D2S346, D2S2289, D2S307, D2S2189, D2S2237, D2S155, D2S325, and D2S2382) in and around the ALS2 locus, we found a maximum two-point LOD score of 2.86 at recombination fraction (θ) 0 for the marker D2S309 and a maximum multipoint LOD score of 3.15 between the markers D2S116 and D2S325 (data not shown). Therefore, we genotyped the 45 individuals from the 10 affected families with a total of 15 markers, including the ALS2 critical region. We obtained a maximum two-point LOD score of 5.87 at θ=0 for the marker D2S309 and a maximum multipoint LOD score of 6.66 between the markers D2S116 and D2S2237 (fig. 2). Haplotype analysis (fig. 1) demonstrated no recombinant event in the region between the markers D2S116 and D2S2237. In addition, the affected patients from the first-degree consanguineous families (families 362 and 283) were homozygous for markers between D2S309 and D2S346, including the ALS2 critical region. These results established that IAHSP is linked to the 2q33-35 region in an interval including the ALS2 critical region.
Figure 2.
Multipoint LOD scores for the ALS2 locus. Markers used, with their distance (in cM), are plotted against multipoint LOD scores. A maximum multipoint LOD score of 6.66 was obtained between the markers D2S116 and D2S2237. The grey bar represents the IAHSP locus, and the black bar represents the ALS2 locus. These loci can be superposed to uncover a region between markers D2S116 and D2S2237.
Mutation Detection
The minimal chromosomal region identified in our families is enriched in transcripts particularly expressed in the motor nervous system (Hadano et al. 2001_b_). To test whether IAHSP is allelic to ALS2 at the ALS2 gene locus, we sequenced the alsin cDNA from EBV-transformed lymphoblasts from one affected patient in each family.
We detected four mutations (in families 362, 419, 283, and 278): three deletions and one splice-site mutation. All the mutations lead to abnormal short and long ALS2 transcripts and truncated alsin proteins (fig. 3).
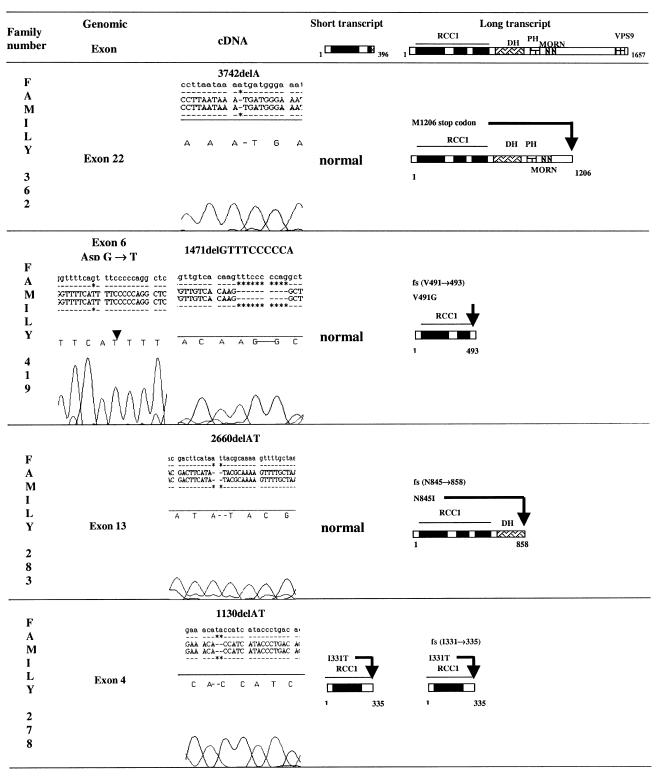
Figure 3.
Electrophoregram results of alsin cDNA sequencing obtained from RT-PCR with RNA extracted from EBV-transformed lymphoblasts of one affected patient from each of the following families: 362, 419, 283, and 278. Three homozygous deletions (in families 362, 283, and 278) were found. All resulted in truncated alsin proteins, involving the short variant only in family 278. For family 419, cDNA analysis demonstrated a 12-bp deletion in the long ALS2 transcript, leading to a truncated long alsin protein. Genomic DNA analysis of the affected patient demonstrated a homozygous point mutation (G→T) in the consensus acceptor splice site (CAG) of exon 6.
In family 362, a single–base pair deletion (3742delA) was detected in exon 22 of the long ALS2 transcript, leading to a stop codon at the 1206 aa position. The three affected patients (patients 2, 3, and 5) were homozygous for the deletion, whereas both parents and one unaffected sibling were heterozygous and an unaffected sibling was homozygous for the wild-type allele (fig. 1).
In family 419, a homozygous 10-bp deletion (1471delGTTTCCCCCA) was detected in exon 6 of the long ALS2 transcript. This deletion leads to a frameshift (V491→) and a premature stop codon at the 493 aa position. Genomic DNA analysis with primers amplifying exon 6 and the corresponding splice-site regions demonstrated a homozygous point mutation (G→T) in the consensus CAG-acceptor splice site of exon 6. Both parents and one unaffected sibling were heterozygous for the mutation (fig. 1).
In family 283, a 2-bp deletion (2660del AT) was detected in exon 13 of the long ALS2 transcript. This deletion leads to a frameshift (N845→), and a premature stop codon 12 codons after the deletion site at the 858 aa position. Genomic DNA analysis of exon 13 demonstrated that the affected patient was homozygous for this deletion and that her parents were heterozygotes, whereas the unaffected brother was homozygous for the normal allele (fig. 1).
In family 278, a homozygous 2-bp deletion (1130delAT) was detected in exon 4 of both the short and the long ALS2 transcripts. The deletion leads to a frameshift (I331→) and a premature translation termination at the 335 aa position. Genomic DNA analysis of exon 4 demonstrated that both parents and the unaffected sibling were heterozygous for the deletion (fig. 1). In the six remaining families (families 279, 408, 242, 751, 747, and 786), no abnormalities were detected by amplification and sequencing of the entire alsin cDNA.
Linkage Analysis in Families without Mutations
Two-point and multipoint LOD scores obtained with the 10 markers of the ALS2 critical region for the six families without mutations were compared with those obtained for the four families with alsin mutations. We found a maximum two-point LOD score of 2.02 at θ=0 for the marker D2S2289. The two markers flanking the ALS2 gene have maximum two-point LOD scores of 1.84 for D2S309 and 0.32 for DS2309 at θ=0 (table 3). A maximum multipoint LOD score of 2.42 between the markers D2S116 and D2S2237 was observed.
Table 3.
Two-point LOD Scores at the Maximum-Likelihood Estimate (_Z_max) of θ between IAHSP and Polymorphic Markers of the ALS2 Locus
| FamilieswithoutMutations(n = 6) | Families withMutations(n = 4) | Total | ||||
|---|---|---|---|---|---|---|
| Locus | _Z_max | θ | _Z_max | θ | _Z_max | θ |
| D2S116 | 1.30 | .00 | 3.82 | .00 | 5.12 | .00 |
| D2S309a | 1.84 | .00 | 4.03 | .00 | 5.87 | .00 |
| D2S2309 | 0.32 | .00 | 2.86 | .00 | 3.18 | .00 |
| D2S2214 | 0.34 | .00 | 3.06 | .00 | 3.40 | .00 |
| D2S346 | 1.84 | .00 | 1.52 | .00 | 3.36 | .00 |
| D2S2289 | 2.02 | .00 | 3.11 | .00 | 5.13 | .00 |
| D2S72 | 1.84 | .00 | 2.92 | .00 | 4.76 | .00 |
| D2S307 | 1.54 | .00 | 2.94 | .00 | 4.48 | .00 |
| D2S2189 | 1.64 | .00 | 3.51 | .00 | 5.15 | .00 |
| D2S2237 | 1.84 | .00 | 3.61 | .00 | 5.45 | .00 |
Discussion
We demonstrated alsin mutations in IAHSP, suggesting that IAHSP is allelic to juvenile ALS and JPLS. IAHSP can be distinguished from those two disorders by the association of (1) an infantile onset, at the age of walking achievement, with an ascending progression during childhood but a long survival during adulthood; (2) absence or severe delay of MEP, contrasting with normal SSEP, EMG, and peripheral NCVs; and (3) a progressive atrophy of the pyramidal tracts apparent on MRI after >20 years of evolution, which suggests a primitive, early degeneration of the upper motor neurons.
The different geographic origins of our families demonstrate that alsin mutations are not restricted to consanguineous Arabic families. The common Mediterranean origin of the three families with mutations we studied and of other reported families with mutations suggests that alsin mutations are more frequent in this region than was initially thought.
Analyzing ALS2 cDNA obtained from lymphoblastoid cell lines, we found four different mutations leading to truncated gene products. However, we did not observe any straightforward genotype-phenotype correlation. Mutations in either the short or the long form of alsin, or in both forms, gave rise to similar phenotypes and disease progression. For instance, in family 278 we detected a mutation that affected both the short and the long alsin transcripts, despite the affected patient's having displayed no signs of lower motor involvement at the age of 24 years. This seems to argue against the hypothesis that an intact short form of alsin protects the lower motor neurons of the spinal cord and brain stem from being affected (Hadano et al. 2001). Short and long alsin transcripts are widely expressed in the brain and spinal cord but also in other tissues outside the CNS (Hadano et al. 2001_a_), including the peripheral sensory nerve (E.E.-P., unpublished data). In addition, only ∼50% of the patients in the Tunisian family with ALS2 originally described showed denervation on EMG analysis (Ben Hamida et al. 1990). Thus, lower motor-neuron involvement does not relate to mutations in the short alsin, as hypothesized elsewhere (Yang et al. 2001). Our results suggest that alsin mutations are responsible for a primitive retrograde degeneration of the upper motor neurons of the pyramidal tracts. In a subgroup of patients, additional unidentified factors might contribute to lower motor-neuron involvement. The large variability in the size of truncated proteins does not correlate with the severity of the phenotype and suggests loss of function. Further studies at the protein level, using alsin antibodies, are certainly needed.
The function of alsin is still unknown, but several homology domains, including RCC1 (guanine exchange factor [GEF] of Ran GTPase [RanGEF]), Db1-pleckstrin (RhoGEF), VPS9 (GEF for GTPase Rab5 vacuolar protein-sorting 21 protein), and two MORN (membrane occupation and recognition plexus) motifs have been found. Alsin is unique in that it might exhibit different potential GEF activities (GEFRho, GEFRan, and GEFRab). GEFs are known to associate with the GDP-bound form of GTPases and to accelerate GDP dissociation and GTP binding, thereby activating the GTPases. This suggests potential functions involving regulation of microtubule assembly, signalling cascades, neuronal morphogenesis, membrane transport or trafficking, organization of the actin cytoskeleton, vacuolar protein sorting, or endocytic trafficking (Hadano et al. 2001_a_; Yang et al. 2001). Alsin can play a key function in neurons as a regulator of the balance between these different GEFs.
Despite a relatively high LOD score without identifiable genetic heterogeneity in the ALS2 genomic critical region, in 6 of the 10 families with IAHSP, no alsin mutations were found by sequence analysis of illegitimate alsin transcripts amplified by RT-PCR from total RNA extracts of lymphoblastoid cell lines. Further analyses are needed to rule out mutations in the intronic and regulatory regions of the ALS2 gene. Quantification of alsin transcripts from tissues with a constitutive expression of the ALS2 gene, easy to collect as peripheral nerve biopsy specimens in affected patients, would be of great interest. An alternative hypothesis would be the presence of a true genetic heterogeneity in IAHSP. Such genetic heterogeneity exists in JPLS; one family with a recombinant event within the ALS2 critical region has been described elsewhere (Yang and al. 2001). In our group of families with IAHSP without identified alsin mutations, no recombination was observed. Surprisingly, however, affected individuals of family 408 are not homozygous for the haplotype cosegregating with the ALS2 locus, despite both parents' having come from the same Libyan tribe, with a high degree of consanguinity. A nonsignificant maximum LOD score of 2.02 was obtained for D2S2289, a marker at a 0.5-cM distance from the ALS2 gene in the ALS2 critical region. Involvement of an another nearby gene in this critical region, rich in transcripts expressed in the motor nervous system (Hadano et al. 2001), cannot be excluded.
We show that mutations in alsin are associated with a subset of the IAHSP phenotype. Thus, alsin mutation is responsible for a primitive, retrograde degeneration of the upper motor neurons of the pyramidal tract, leading to a clinical continuum from infantile (IAHSP) to juvenile forms with (ALS2) or without (JPLS) lower motor-neuron involvement. Further analyses will determine whether other hereditary disorders characterized by primary degeneration of the central motor pathways, such as pure forms of spastic paraplegia, could be the result of alsin dysfunction.
Acknowledgments
The patients and their families are warmly acknowledged for their participation. We greatly thank L. Dauche, F. Gauthier, and G. Giraud for their help in processing blood samples and in genotyping. This work was supported by grants from the European Leukodystrophy Association, INSERM (projet PROGRES), and the Jean Pierre and Nancy Boespflug’ Myopathic Research Foundation. We are also indebted for a grant from the Italian Ministry of Health for Ricerca Finalizzata Strategica on genetic leukodystrophies and spastic paraplegia.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://www.marshfieldclinic.org/research/genetics/ (for order of and distance between markers)
- Centre de Ressources INFOBIOGEN, http://www.infobiogen.fr/ (for linkage analysis support)
- Généthon, http://www.genethon.fr/php/index.php (for primer sequences)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for spastin [MIM 604277], paraplegin [MIM 604277], L1CAM [MIM 308840], PLP [MIM 312080], atlastin [MIM 606439], HSP90 [MIM 118190], SOD1 [MIM 147450], alsin [MIM 606352], SPG1 [MIM 312900], SPG2 [MIM 312920], SPG3A [MIM182600], SPG4 [MIM182601], SPG7 [MIM 602783], SPG13 [MIM 605280], ALS1 [MIM105400], ALS2 [MIM 205100], ALS4 [MIM 602433], ALS5 [MIM 602099], ALS6 [MIM 606640], and PLSJ [MIM 606353])
- Web Resources of Genetic Linkage Analysis, http://linkage.rockefeller.edu/ (for LINKAGE package, version 5.1, generation of pedigree file [MAKEPED] and data file [PREPLINK], using the allele frequency of each marker found in CEPH database)
References
- Ben Hamida M, Hentati F, Ben Hamida C (1990) Hereditary motor system diseases (chronic juvenile amyotrophic lateral sclerosis). Brain 113:347–363 [DOI] [PubMed] [Google Scholar]
- Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A (1998) Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 93:973–983 [DOI] [PubMed] [Google Scholar]
- Chance PF, Rabin BA, Ryan SG, Ding Y, Scavina M, Crain B, Griffin JW, Comblath DR (1998) Linkage of the gene for autosomal dominant form of juvenile amyotrophic lateral sclerosis to chromosome 9q34. Am J Hum Genet 62:633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figlewicz DA, Bird TD (1999) “Pure” hereditary spastic paraplegias: the story becomes complicated. Neurology 53:5–7 [DOI] [PubMed] [Google Scholar]
- Hadano S, Hand C, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH Jr, Scherer SW, Rouleau GA, Hayden MR, Ikeda JE (2001_a_) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29:166–173 [DOI] [PubMed] [Google Scholar]
- Hadano S, Nichol K, Brinkman RR, Nasir J, Martindale D, Koop BF, Nicholson DW, Scherer SW, Ikeda JE, Hayden MR (1999) A yeast artificial chromosome–based physical map of the juvenile amyotrophic lateral sclerosis (ALS2) critical region on human chromosome 2q33-q34. Genomics 55:106–112 [DOI] [PubMed] [Google Scholar]
- Hadano S, Yanagisawa Y, Skaug J, Fichter K, Nasir J, Martindale D, Koop BF, Scherer SW, Nicholson DW, Rouleau GA, Ikeda J, Hayden MR (2001_b_) Cloning and characterization of three novel genes, ALS2CR1, ALS2CR2, and ALS2CR3, in the juvenile amyotrophic lateral sclerosis (ALS2) critical region at chromosome 2q33-q34: candidate genes for ALS2. Genomics 71:200–213 [DOI] [PubMed] [Google Scholar]
- Hand CK, Khoris J, Salachas F Gros-Louis F, Lopes AA, Mayeux-Portas V, Brown RH Jr, Meininger V, Camu W, Rouleau GA (2002) A novel locus for familial amyotrophic lateral sclerosis on chromosome 18q. Am J Hum Genet 70:251–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JJ, Dürr A, Cournu-Rebeix I, Georgopoulos C, Ang D, Nielsen MN, Davoine CS, Brice A, Fontaine B, Gregersen N, Bross P (2002) Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet 70:1328–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding AE (1981) Hereditary “pure” spastic paraplegia: a clinical and genetic study of 22 families. J Neurol Nurosurg Psychiatry 44:871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine CS, Cruaud C, Durr A, Wincker P, Brottier P, Cattolico L, Barbe V, Burgunder JM, Prud'homme JF, Brice A, Fontaine B, Heilig B, Weissenbach J (1999) Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet 23:296–303 [DOI] [PubMed] [Google Scholar]
- Hentati A, Bejaoui K, Pericak-Vance MA, Hentati F, Speer MC, Hung WY, Figlewicz DA, Haines J, Rimmler J, Ben Hamida C, et al (1994) Linkage of recessive familial amyotrophic lateral sclerosis to chromosome 2q33-q35. Nat Genet 7:425–428 [DOI] [PubMed] [Google Scholar]
- Hentati A, Ouahchi K, Pericak-Vance MA, Nijhawan D, Ahmad A, Yang Y, Rimmler J, Hung W, Schlotter B, Ahmed A, Ben Hamida M, Hentati F, Siddique T (1998) Linkage of a commoner form of recessive amyotrophic lateral sclerosis to chromosome 15q15-22 markers. Neurogenetics 2:55–60 [DOI] [PubMed] [Google Scholar]
- Jouet M, Rosenthal A, Armstrong G, MacFarlane J, Stevenson R, Paterson J, Metzenberg A, Ionasescu V, Temple K, Kenwrick S (1994) X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat Genet 7:402–407 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1985) Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet 37:482–498 [PMC free article] [PubMed] [Google Scholar]
- McDermott C, White K, Bushby K, Shaw P (2000) Hereditary spastic paraparesis: a review of new developments. J Neurol Neurosurg Psychiatry 69:150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder DW, Kurland LT, Offord KP, Beard CM (1986) Familial adult motor neuron disease: amyotrophic lateral sclerosis. Neurology 36:511–517 [DOI] [PubMed] [Google Scholar]
- Pringle CE, Hudson AJ, Munoz DG, Kiernan JA, Brown WF, Ebers GC (1992) Primary lateral sclerosis: clinical features, neuropathology and diagnostic criteria. Brain 115:495–520 [DOI] [PubMed] [Google Scholar]
- Reid E, Dearlove AM, Osborn O, Rogers MT, Rubinsztein DC (2000) A locus for autosomal dominant “pure” hereditary spastic paraplegia maps to chromosome 9q13. Am J Hum Genet 66:728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al (1993) Mutations in Cu/Zn superoxidase dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62 [DOI] [PubMed] [Google Scholar]
- Saugier-Veber P, Munnich A, Bonneau D, Rozet JM, Le Merrer M, Gil R, Boespflug-Tanguy O (1994) X-linked spastic paraplegia and Pelizaeus-Merzbacher disease are allelic disorders at the proteolipid protein locus. Nat Genet 6:257–262 [DOI] [PubMed] [Google Scholar]
- Seri M, Cusano R, Forabosco P, Cinti R, Caroli F, Picco P, Bini R, Morra VB, De Michele G, Lerone M, Silengo M, Pela I, Borrone C, Romeo G, Devoto M (1999) Genetic mapping to 10q23.3-q24.2, in a large Italian pedigree, of a new syndrome showing bilateral cataracts, gastroesophageal reflux, and spastic paraparesis with amyotrophy. Am J Hum Genet 64:586–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique T, Figlewicz DA, Pericak-Vance MA, Haines JL, Rouleau G, Jeffers AJ, Sapp P, Hung WY, Bebout J, McKenna-Yasek D, et al (1991) Linkage of a gene causing familial amyotrophic lateral sclerosis to chromosome 21 and evidence of genetic-locus heterogeneity. N Engl J Med 324:1381–1384 [DOI] [PubMed] [Google Scholar]
- Vazza G, Zortea M, Boaretto F, Micaglio GF, Sartori V, Mostacciuolo ML (2000) A new locus for autosomal recessive spastic paraplegia associated with mental retardation and distal motor neuropathy, SPG14, maps to chromosome 3q27-q28. Am J Hum Genet 67:504–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, Cole N, Gascon G, Yagmour A, Ben-Hamida M, Pericak-Vance M, Hentati F, Siddique T (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29:160–165 [DOI] [PubMed] [Google Scholar]
- Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, Tukel T, Apak M, Heiman-Patterson T, Ming L, Bui M, Fink JK (2001) Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet 29:326-331 [DOI] [PubMed] [Google Scholar]