Transcription Factors in Long-Term Memory and Synaptic Plasticity (original) (raw)
. Author manuscript; available in PMC: 2014 Jan 7.
Published in final edited form as: Physiol Rev. 2009 Jan;89(1):10.1152/physrev.00017.2008. doi: 10.1152/physrev.00017.2008
Abstract
Transcription is a molecular requisite for long-term synaptic plasticity and long-term memory formation. Thus, in the last several years, one main interest of molecular neuroscience has been the identification of families of transcription factors that are involved in both of these processes. Transcription is a highly regulated process that involves the combined interaction and function of chromatin and many other proteins, some of which are essential for the basal process of transcription, while others control the selective activation or repression of specific genes. These regulated interactions ultimately allow a sophisticated response to multiple environmental conditions, as well as control of spatial and temporal differences in gene expression. Evidence based on correlative changes in expression, genetic mutations, and targeted molecular inhibition of gene expression have shed light on the function of transcription in both synaptic plasticity and memory formation. This review provides a brief overview of experimental work showing that several families of transcription factors, including CREB, C/EBP, Egr, AP-1, and Rel have essential functions in both processes. The results of this work suggest that patterns of transcription regulation represent the molecular signatures of long-term synaptic changes and memory formation.
I. Introduction
Memory, the capacity to retain learned information, can be divided on the basis of its duration, into short- and long-term (223). Although several intermediate states are less well understood, short-term and long-term memories are accompanied by different biological states and mechanisms of retention and therefore are two distinct forms of memory. A short-term memory is the retention of information for a brief time without creation of the neural changes for later recall (e.g., obtaining and using a phone number from directory assistance). In contrast, a long-term memory occurs when, following learning, changes in neural pathways take place for the storage of information that can be recalled weeks, months, or even years later. An important biological feature distinguishing short- from long-term memory is that only the latter depends on a temporally limited phase of RNA and protein synthesis. If either RNA or protein synthesis is blocked before or immediately after training, long-term memory formation is disrupted (53). In a great number of studies done over the last 50 years, translational inhibitors have been used to characterize the requirement for protein synthesis during memory formation (14, 78, 91, 96, 122, 227). Although inhibitors of transcription such as actinomycin D have been used less frequently, they have shown, in several learning tasks and different species, that transcription, like translation, is an essential step for memory formation.
Newly learned information exists for a limited time in a labile state. With the passage of time, however, this information becomes stable and insensitive to disruption, a process known as memory consolidation (153). During the initial phase of consolidation, memory formation can be prevented or disrupted by various interferences, including additional learning, seizure, trauma, brain cooling, neuronal inactivation, brain lesions or inactivation, inhibition of transcription or specific transcription factors, and inhibition of translation or selective blockade of certain molecular pathways. Once memory has stabilized, it is not forever insensitive to disruption but can again become temporally labile if reactivated, for example, by recall. During this new phase of fragility, as during the initial post-training consolidation phase, memory can be disrupted if transcription or translation is inhibited. Because similar interferences can affect the stability of memory after initial learning and recall, the process that transforms a reactivated memory from a labile to a stable form is called reconsolidation (12, 61, 180, 213). Hence, given that transcription is an essential step for both memory consolidation and reconsolidation, how does it contribute to the changes underlying the whole process of memory formation? Which transcription factors are involved? What genes are ultimately regulated and required to mediate memory formation? How is transcription regulated over time? Can we change the pattern of transcription and regulate the intensity of memory retention? Such questions have been the focus of many studies, which, since the 1960s, have attempted to elucidate the biological bases underlying memory formation. Here I will summarize current knowledge about the function of transcription factor families and their regulation during memory consolidation and reconsolidation.
IA. Inhibitors of Transcription and Memory
As noted earlier, the use of transcriptional inhibitors in studies of memory has not been as widespread as the use of translational inhibitors, mostly due to their toxicity and side effects (198, 250). Nevertheless, investigations based on the use of these compounds have suggested that a critical time window of transcription is necessary for the formation of long-term memory. Indeed, studies based on the use of actinomycin D have indicated, in species ranging from sea snails and goldfish to monkeys, that inhibiting mRNA transcription around the time of training blocks long-term memory retention (4, 39, 182, 192, 222, 235, 250).
Recent studies have shown that multiple waves of protein synthesis occur after learning or long-term synaptic plasticity (2, 27). In agreement with this finding, more than one phase of transcription has been shown to be required for memory formation. For example, injections into the CA1 regions of the rat hippocampus (a region required for spatial and contextual types of memories) of either the adenosine analog DRB or the RNA polymerase II inhibitor alpha-amanitin showed that at least two periods of transcription are needed to establish a long-term inhibitory avoidance memory, the first at about the time of training and the second 3–6 hr later. These periods seem to overlap with those needed for translation, suggesting that the multiple phases of transcription are indeed coupled to translation (101, 197). The same transcriptional blockers have been stereotactically injected into the hippocampus of rats after either training or retrieval of a spatial memory. This treatment blocked not only the initial consolidation, but also the postretrieval reconsolidation of memory, suggesting that both processes are dependent on transcription (51). Similar results on reconsolidation have been found in the invertebrate Lymnaea stagnalis (211). In this species, either post-retrieval administration of actinomycin D or ablation of the soma of a specific neuron known to mediate consolidation of a conditioned response disrupts reconsolidation. However, the necessity of transcription in memory reconsolidation has not been confirmed in all cases. For example, injection of actinomycin D into the amygdala after recall of a fear memory had no effect although, with the same paradigm, the injection of translational inhibitors disrupted memory at subsequent tests (191).
Long-term synaptic plasticity responses such as long-term facilitation (LTF) in the invertebrate _Aplysia californica_as well as long-term potentiation (LTP) and long-term depression (LTD) in the mammalian brain, are like memory in that they require transcription, wheras short-term responses do not (44, 71). Long-term synaptic plasticity responses are long-lasting changes in the strength of synaptic connections and are believed to represent the cellular correlate of learning and memory formation (145).
Despite some inconsistent findings, which may be due to differences in behavioral or molecular experimental conditions, a multitude of studies, as I will detail, have identified various transcription factors that have a crucial function during either the initial consolidation or the reconsolidation of several types of memories in many different species. It should be kept in mind that memory experiments are carried out in several species using many different memory tasks and experimental designs. Consequently, some contrasting results may be explained by these differences.
IB. Transcription Factors and Transcription Regulation
The genetic information carried by all organisms is expressed in any biological function via a series of processes that begins with DNA transcription. Through this process, the information encoded in the DNA of each cell is copied into a molecule of RNA. This occurs through three steps. The first step, _initiation_consists of binding of the enzyme RNA polymerase to double-stranded DNA, which becomes single-stranded in the region of the binding; the region of DNA bound by the RNA polymerase, together with the general transcription factors necessary for transcription, is the promoter. The second step, _elongation_involves the covalent addition of nucleotides to the 3' end of the growing polynucleotide chain. The third step, _termination_is the recognition of the transcription termination sequence and the release of RNA polymerase (15).
RNA polymerase is the enzyme that mediates transcription. However, in eukaryotic cells, as demonstrated by Roeder and colleagues (204), this enzyme alone does not function efficiently; it needs to interact with other proteins, known as transcription factors, to produce transcripts (Fig. 1). These factors either interact directly with the RNA polymerase or regulate its catalytic function by binding to cis-acting DNA sequences or another transcription factor. The RNA polymerase and the transcription factors that directly interact with it are the basal transcription machinery. This apparatus is directly responsible for transcription. On its own, however, the basal transcription machinery is not efficiently regulated, so that it, in turn, requires additional transcription factors to either accelerate or repress transcription; these are known as upstream factors. The upstream factors regulate the efficiency of transcription by influencing different transcription steps. For example, the upstream factors regulate the initiation step through interactions with members of the basal transcription apparatus, promoting assembly of the basal apparatus and/or binding cofactors that interact with the basal apparatus. The function and expression of some transcription factors are temporally or spatially regulated and/or occur directly in response to the environment, which is the final link in controlling gene expression. These transcription factors are inducible transcription factors. The RNA polymerase of interest in this review is the RNA polymerase II, which transcribes protein-encoding genes into messenger RNAs (mRNAs), as well as sequences encoding small nuclear RNAs.
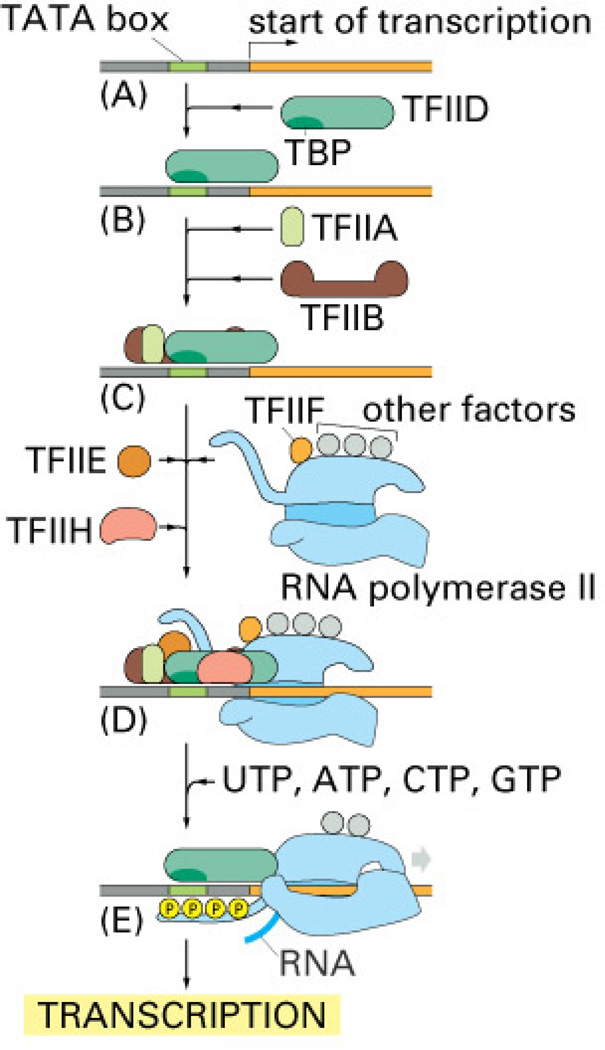
Figure 1. Transcription.
To begin transcription, eucaryotic RNA polymerase II requires the general transcription factors. These transcription factors are called TFIIA, TFIIB, and so on. (A) The promoter contains a DNA sequence called the TATA box, which is located 25 nucleotides away from the site where transcription is initiated. (B) The TATA box is recognized and bound by transcription factor TFIID, which then enables the adjacent binding of TFIIB. (C) For simplicity the DNA distortion produced by the binding of TFIID is not shown. (D) The rest of the general transcription factors, as well as the RNA polymerase itself, assemble at the promoter. (E) TFIIH uses ATP to pry apart the double helix at the transcription start point, allowing transcription to begin. TFIIH also phosphorylates RNA polymerase II, releasing it from the general factors so it can begin the elongation phase of transcription. As shown, the site of phosphorylation is a long polypeptide tail that extends from the polymerase molecule. The exact order in which the general transcription factors assemble on each promoter is not known with certainty. In some cases, most of the general factors are thought to first assemble with the polymerase independent of the DNA, with this whole assembly then binding to the DNA in a single step. The general transcription factors have been highly conserved in evolution; some of those from human cells can be replaced in biochemical experiments by the corresponding factors from simple yeasts. [From Alberts et al. (16].
In eukaryotic cells, general transcription factors are remarkably conserved throughout evolution. They include the TATA box binding protein (TBP) and TBP-associated factors (TAFs), which together form transcription factor IID (TFIID), as well as the transcription factors IIB, IIE, IIF, IIH and IIA(TFIIB, TFIIE, TFIIF TFIIH and TFIIA, (Fig. 1) (for reviews, see 86, 187, 205, 267). Thus, a complex of general transcription factors, together with upstream factors, which can be either constitutively expressed or inducible, cooperate in the regulation of transcription of each gene. Like the TBP that recognizes the DNA sequence known as TATA box, other transcription factors interact directly with DNA by recognizing specific DNA sequence motifs, or DNA binding sites. The transcription factors bind to these sequences within the major grove of DNA via α helices or β sheets, structures that serve as DNA binding domains. Once the transcription factors are bound to these domains, their regulatory domains can regulate transcription. Thus, concerted action by a multitude of transcription factors that bind to DNA regulatory regions determines which of its thousands of genes a cell will transcribe at a given time. Some regulatory regions are simple, acting as switches that are thrown by a single signal; many others are complex, acting as tiny microprocessors that respond to a variety of signals, which they interpret and integrate to switch their target genes on or off.
Many functions are common to all cells, and therefore rely on the expression of common proteins. including the structural proteins of chromosomes, RNA polymerases, DNA repair enzymes, ribosomal proteins, enzymes involved in the central reactions of metabolism, and proteins that form the cytoskeleton. Other functions are specialized for given types of cells. For example, immunoglobulins are found only in lymphocytes and plasma cells, while certain neurotransmitter receptors are expressed only in neurons.
Investigations of mRNA transcripts suggest that, at any one time, a typical human cell expresses approximately 10,000–20,000 of its approximately 30,000 genes. The level of expression of every active gene generally varies from one cell type to another. A few of these differential expressions are striking, like that of immunoglubulins noted earlier. However, most are much more subtle. Nevertheless, the patterns of mRNAs in a given cell are not only characteristic of the cell type, but change in response to environmental challenges. In fact, most eukaryotic cells are capable of altering their patterns of gene expression in response to extracellular stimuli. Moreover, different cell types often respond in different ways to the same extracellular stimulus. It recently was demonstrated that individual neuronal cell types have highly variable levels of protein expression, including that of ion channels (215). In addition, a taxonomic tree constructed using microarray analyses of 12 distinct populations of neurons from different regions or different subpopulations within each region of the adult mouse forebrain indicated highly heterogeneous gene expression even within a single region (226).
In summary, transcription is the first regulatory step in gene expression. It is mediated by the concerted action of a multitude of basal and regulated transcription factors that, at a given time, come together to mediate the transcription of specific genes. Thus, transcription represents the molecular signature of the expression of each gene at a given time in the context of each cell, providing an interesting example of the uniqueness of each cell type’s regulation of its functional response to specific environmental stimuli over time.
II. Families of Transcription Factors Involved in Synaptic Plasticity and Memory Formation
Later than those in many other fields such as immunology, cell biology, developmental biology, and cancer biology, researchers in neuroscience have begun to investigate the functions of transcription and its regulation in specific cells and tissues. In the last 15 years, many studies have focused on unraveling the nature and function of gene expression in the brain during its development, as well as during behavioral responses in adulthood. Despite this effort, a great deal remains unknown. For example, one issue of particular interest is understanding the transcription regulation underlying cognitive functions. How does gene expression allow complex cognitive responses? What types of genes mediate such “evolved” functions? What is the cellular result of the expression of those genes?
Studies based on genome sequences of several species and their comparative analyses have revealed surprising simplicity in the evolution of the genetic content. Notably, the number of genes in the genome of the different species does not account for the evolutionary increase in morphological and behavioral complexity. Indeed, while the genome in C. elegans contains approximately 20,000 genes, the number of protein-coding genes in the human genome is only about 30,000. Moreover, it seems that this small increase is primarily due to duplication of existing genes rather than the creation of new ones. Given this simplicity, how is evolutionary complexity generated? If the human genome is not very different from that of a worm, how is the complexity of human behavior accounted for?
One interesting hypothesis proposes that behavioral complexity correlates with the number of patterns of gene expression that an organism produces (140). This hypothesis is supported by the fact that 5%-10% of the total coding capacity of a metazoan genome is made up of transcription factors, and the human genome contains as many as 3,000 transcription factors. Considering that the combinatorial nature of transcription regulation produces remarkable amplification of the repertoire of gene expression, and that the brain is the tissue in which changes in protein and gene expression are most pronounced, it is strongly tempting to speculate that the biological bases of cognition lie within the concerted action of a complex regulation of gene expression.
In this review I will focus on the families of transcription factors thus far found to be critically involved in synaptic plasticity and memory formation. These include cAMP response element binding protein (CREB), CCAAT enhancer binding protein (C/EBP), activating protein 1 (AP-1), early growth response factor (Egr), and Rel/Nuclear Factor kB (Rel/NF-kB). It is interesting to note that, at least according to current knowledge, most transcription factor families that are implicated in synaptic plasticity and in learning and memory in the adult brain are distinct from those found to be primarily involved in the development of the nervous system, including Hes, paired domain, paired homeodomain, LIM homeodomain, RE-1 silencing transcription factor, GATA, and SOX genes (233). Future studies should reveal whether this reflects a biological functional dissociation or a lack of overlapping investigations between development and cognition.
IIA. cAMP Response Element Binding Protein (CREB) and the Evolutionarily Conserved CREB-Dependent Gene Expression Required for Long-Term Synaptic Plasticity and Memory Formation
In the late 80s, it was discovered that cyclic AMP (cAMP) mediates the hormonal stimulation of several cellular processes by regulating the phosphorylation of critical proteins, among them the transcription factor cAMP response element binding protein (CREB) (175). Indeed, one important role of protein kinases and phosphatases is to regulate the activities of DNA-binding proteins (transcription factors) that interact with specific DNA binding elements in the promoter regions of genes, thereby activating or repressing their transcription. The classical sequences that are known to be specifically recognized by CREB are the palindromic octanucleotide sequences TGACGTCA, which are referred to as cAMP response elements (CRE) (165, 174).
The CREB (or CREB/ATF) family of transcription factors includes three homologous genes (Fig. 2), creb, cAMP response element modulator (crem), and activating transcription factor-1 (atf-1). These genes generate a group of highly homologous proteins that have been named after their prototypes: CREB, CREM, and ATF-1, respectively (176). While both CREBs and ATF-1 are ubiquitously expressed proteins, CREMs are mainly present in the neuroendocrine system. The cloning of several transcription factors belonging to the CREB family, as well as to the AP-1 and C/EBP families, together with analyses of their predicted encoded proteins, have demonstrated that these families of transcription factors have a common structural motif, the basic region-leucine zipper (bZIP) domain, which generally resides at the C-terminus.
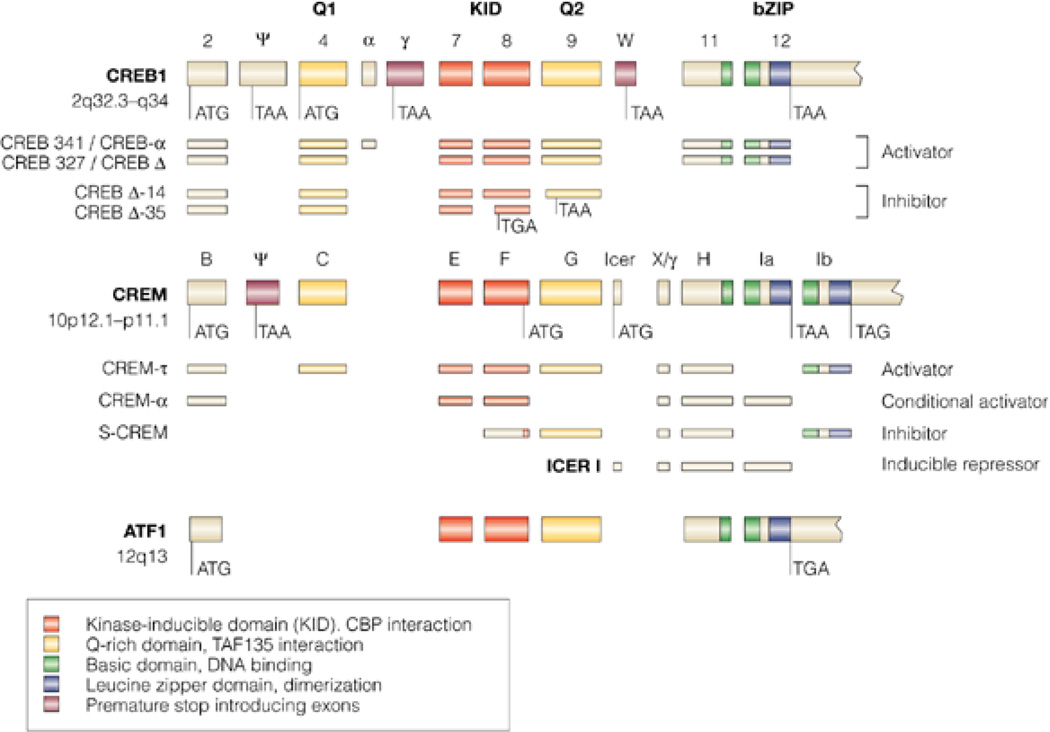
Figure 2. Genomic organization of the mammalian CREB family.
The consensus alignment of the genomic organization of human cyclic AMP response element (CRE)-binding protein (CREB), the cAMP response element modulator (CREM) and the activating transcription factor 1 (ATF1) was obtained by BLAST analysis of human GenBank sequences against the human genome and against each other. In cases for which human sequences were unavailable for published isoforms, the mouse or rat sequences were used. Homologous domains are in color (see key). In the CREB and CREM genes, several alternatively spliced exons exist, creating in-frame stop codons (TAA or TGA) leading to carboxy-terminally truncated proteins or giving rise to amino-terminally truncated proteins by using internal transcription-initiation sites downstream of the stop-introducing exon. A partial list of alternative splice products with divergent activating properties is shown. [From Mayr & Montminy (151)].
The bZIP domain critically mediates binding to specific regulatory sequences, whereas the rest of the protein, at the N-terminus, contains the transcriptional activation domain. The name bZIP describes two juxtaposed regions: the basic region is rich in basic amino acids; the other contains leucine residues localized at every seven positions so that, through an alpha-helix structure, they create a hydrophobic leucine-enriched surface that mediates dimerization by resembling a zipper (leucine zipper). Through this structure, proteins of the bZIP family bind as dimers to their DNA binding sequences. The sequence of each bZIP domain also governs whether these proteins form homo-dimers or hetero-dimers.
Heterodimerization can occur between CREB family members, but also with other bZIP transcription factors such as C/EBPs, Fos, and Jun proteins. This is important because it generates a great expansion of the repertoire and diversity in the regulation of target genes (83, 151). Moreover, alternative splicing of the CREB and CREM genes contributes to the vast repertoire of CREB family members, which include several isoforms that have distinct repressor or activator features (151). The transactivation domain of CREB is bipartite, including a constitutive (Q2) and an inducible domain, the kinase-inducible domain (KID). The Q2 domain mediates interaction with a component of the TFIID complex, whereas the KID seems to promote isomerization by recruiting the co-activator factors CREB binding protein (CBP) and p300 to the promoters. The KID region is active only when it is phosphorylated at Ser 133, which occurs in response to cAMP and is critical for the activation of CREB (47, 151). CBP and p300 are transcription co-activators with intrinsic histone acetyltransferase activity, which is necessary for mediating transcription enhancement (107, 112). Several reviews have described the structural and biological features of CREB CREM and ATF-1 in greater detail (55, 143, 151, 176).
CREB factors are critical in many important functions in the nervous system, including neurogenesis and neuronal survival, development, and differentiation, as well as neuroprotection, axonal outgrowth and regeneration, circadian rhythms, addiction, synaptic plasticity, and memory formation (24, 43, 60, 171, 177, 194, 219).
CREB is known to be generally constitutively expressed and to be activated via posttranslational modifications such as phosphorylation. Phosphorylation of its residue Ser 133 is particularly important because it mediates activation of the KID domain. The phosphorylation of CREB can be triggered by a variety of signaling processes, including an increase in intracellular Ca++ via activation of voltage- or ligand-gated channels such as NMDA receptors, an increase in cAMP via activation of G-coupled receptors, or activation of receptor tyrosine kinase by growth factors (143). Thus, CREB is a central activator that can integrate a variety of stimuli (Fig. 3).
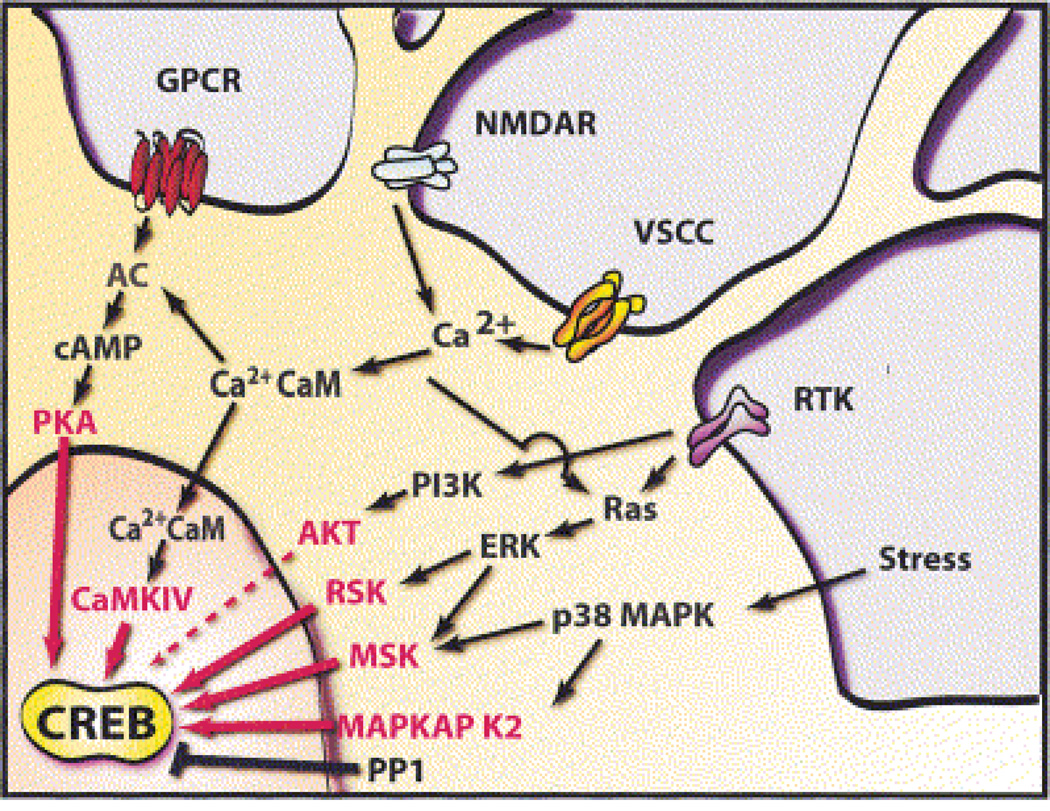
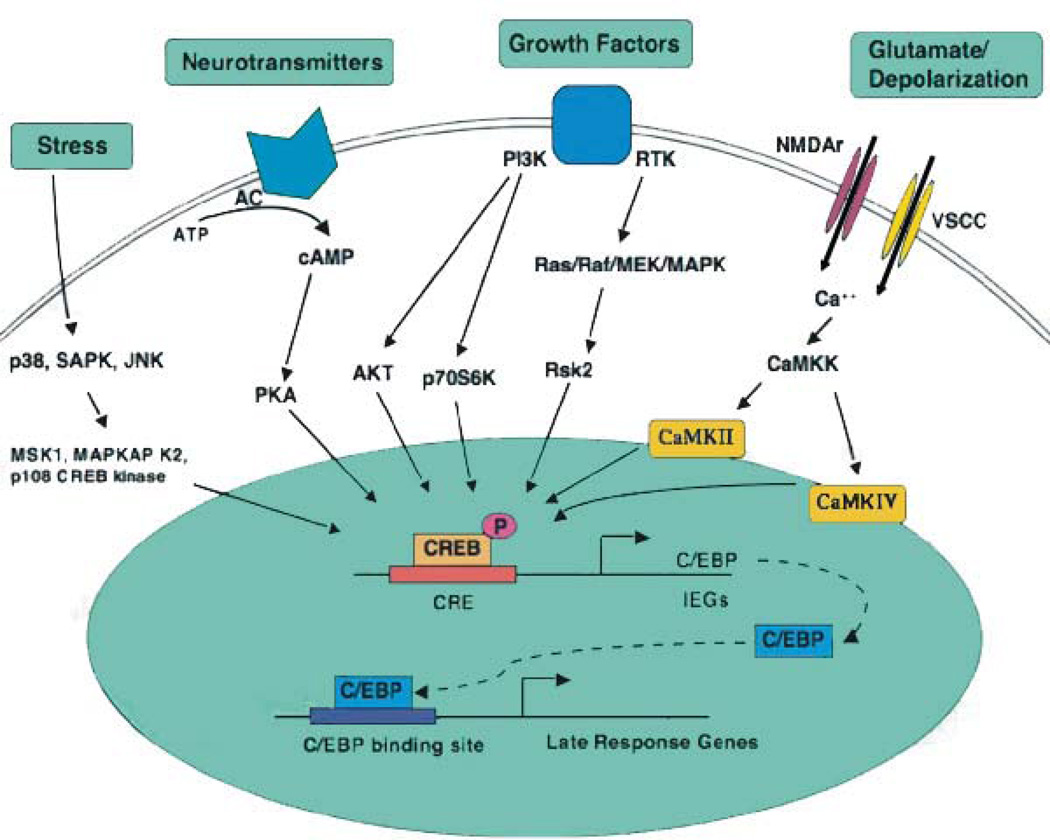
Figure 3. An overview of signaling pathways that converge on CREB.
Excitatory neurotransmitters, ligands for GPCRs, neuronal growth factors, and stress inducers are among the stimuli that activate signaling pathways that converge upon CREB. Multiple stimulus-dependent protein kinases have been implicated as CREB kinases in neurons, and a high degree of crosstalk exists between these signaling pathways. Stimulus-dependent CREB kinases include PKA, CaMKIV, MAPKAP K2, and members of the pp90RSK (RSK) and MSK families of protein kinases. Protein phosphatase 1 (PP1) has been implicated as the predominant phospho-CREB phosphatase. [From Lonze and Ginty (143)].
In the 1980s, studies of the invertebrate Aplysia californica advanced our knowledge of the molecular changes underlying short- and long-term memory. A simple form of learning and memory in Aplysia is sensitization of the gill and siphon withdrawal reflex in which the animal learns about a noxious or aversive stimulus such as a tail shock and consequently enhances its defensive behavior in response to subsequent neutral stimuli. The animal’s defensive behavioral response is withdrawal of its siphon and gill. The duration of this response reflects memory retention. Kandel and colleagues (116) found that the intensity and duration of this withdrawal is a function of the number of shocks received. Whereas the short-term sensitization induced by a single shock lasts for a few minutes and depends on post-translational modifications, multiple shocks induce sensitization that lasts for weeks, requiring new protein and RNA synthesis. Short- and long-term sensitization are, respectively, accompanied by short- and long-term synaptic facilitation of neurotransmitter release.
Castellucci et al. (44) and Montarolo et al. (173) reproduced both short- and long-term facilitation in sensory-motor co-cultures using applications of serotonin, a neurotransmitter found to mediate sensitization. A single serotonin application to the culture produces a facilitation that last only a few minutes and requires post-translational modifications, while repeated applications of serotonin give facilitation that lasts for days and requires new RNA and protein synthesis. Both short and long-term facilitation are mediated by the activation of cAMP and the cAMP-protein kinase A (PKA) signaling pathway. However, only long-term facilitation depends on transcription and translation. The gene expression requirement has been found in several species and in many different learning paradigms, suggesting that it represents an evolutionarily conserved feature of memory formation.
In 1990, based on their use of Aplysia sensory and motor neuron co-cultures, Dash et al. (52) asked whether the gene expression required for long-term facilitation critically recruits cAMP-induced gene regulation? In other words, do CRE-regulated target genes have a critical function in long-term synaptic plasticity and long-term memory? They found that injection of CRE binding sites into sensory neurons, which compete for endogenous CRE binding, disrupts long-term facilitation (LTF) induced by repeated pulses of serotonin (5 hydroxytriptamine, 5-HT) while leaving intact the short-term facilitation (STF) induced by a single pulse of 5HT. This study provided the first demonstration that CRE-dependent gene expression is critical for long-term facilitation but does not affect short-term facilitation. A few years later, the cloning of CREB isoforms in Aplysia californica directly proved the action of CREBs in long-term facilitation (25). Together, these and numerous other studies of Aplysia led to the conclusion that the cAMP-protein kinase A (PKA)-CREB is a critical molecular pathway underlying long-term facilitation (118). Moreover, it was found that not only PKA, but also mitogen-activated protein kinase (MAPK or ERK) is essential in activation of the CREB pathway. In fact, it has been proposed that in the Aplysia model, after 5 pulses of 5HT, activated PKA and MAP kinases translocate to the nucleus, where they presumably phosphorylate an activator form of CREB, CREB1, leading to its activation (148, 166). In parallel, a repressor form of CREB, CREB2, is removed, thus allowing gene expression activation (26).
At the same time, genetic studies of another invertebrate, _Drosophila melanogaster_revealed that mutants with defects in either short-term or long-term memory carried mutations in genes encoding for enzymes involved in activation of the cAMP-PKA-dependent pathway, including adenylyl cyclase, the catalytic subunit of PKA, and cAMP-specific phosphodiesterase. Genetic mutations of the CREB gene pointed to the existence of both activator and inhibitor isoforms of CREBs (262). Although the existence of the activator form was then reported to be controversial (193), the findings in both Aplysia and Drosophila led to the question of whether the CREB-dependent pathway plays an evolutionarily conserved function in long-term memory formation. Thus, the CREB isoforms most abundantly expressed in the brain, the α and Δ isoforms, were knocked out in mice (CREBα/Δ), and these mice were analyzed behaviorally and electrophysiologically. The CREBα/Δ knockout mice showed significant deficits in several types of memories, including spatial, contextual, and cued (34). Subsequent studies confirmed and extended these findings, supporting the conclusion that CREB plays an essential role during memory formation in numerous additional species, brain regions, and learning systems (38, 50, 62, 80, 100, 110, 125, 129, 196, 244, 265, 268).
The CREBα/Δ knock-out mice also showed impaired hippocampal long-term potentiation (LTP) (34). LTP, as noted earlier, is a form of long-term synaptic plasticity believed to represent a possible cellular mechanism for learning and memory (184, 218). Like memory formation, LTP has been subdivided into temporal phases. The early phase (E-LTP), which is believed to be independent from new protein synthesis, involves only post-translational mechanisms and decays within about an hour under high-frequency stimulation (HFS). The protein synthesis-dependent phase (late or L-LTP) begins as early as 15 min after HFS and can persist for several hours (70, 188, 241). It has been proposed that this phase models the gene-expression-dependent phase of long-term memory consolidation (72).
Many investigators confirmed that LTP requires the function of CREBs (23, 30, 102, 111, 217), leading to the conclusion that CREB is a critical mediator of long-term synaptic plasticity and long-term memory formation. However, some authors reported that the essential function of CREB in memory formation may be overcome by genetic backgrounds and that CREB-independent forms of long-term plasticity and memory exist. For example, Balschun et al. (22) found that conditional disruption of all CREB isoforms in the hippocampal CA1 subregion and other forebrain regions is not sufficient to impair hippocampal LTP and LTD as well as contextual fear conditioning, but instead causes only modest impairment in the early stages of water-maze learning. The same authors reported that conditional disruption of all CREB isoforms throughout the brain had no effect on hippocampal LTP and LTD or contextual fear conditioning, although it significantly impaired conditioned taste aversion. Conversely, Pittenger et al. (196), using another conditional mouse line in which the function of all CREB subfamilies was blocked in CA1 neurons of the dorsal hippocampus, reported that although several forms of late-phase LTP were normal, forskolin-induced and dopamine-regulated potentiation were disrupted. These contrasting findings do not confound the conclusion that CREB is critical in LTP and memory formation; in fact, they can be explained by the fact that, in some knock-out lines or learning conditions, protein compensation overcomes the requirement for CREB and/or that some forms of memories and LTP are CREB-independent. Hence, various results indicate that CREB is critical for memory formation.
For how long and in which brain regions is CREB critically involved in memory formation? Answering this question requires that CREB be inactivated or disrupted in a specific region at given and restricted times. This can be achieved, for example, by stereotactic delivery of antisense oligodeoxynucleotides (ODN), RNA interference (RNAi), or viral expression. In 1997, Guzowski and McGaugh (80) injected CREB ODN antisense bilaterally into the rat dorsal hippocampus and showed that in this region CREB-mediated transcription is a necessary component for learning and remembering a spatial task. This requirement is temporally limited to the post-training phase. In fact, the requirement was no longer seen one day after training, suggesting that CREB-dependent gene expression required for memory consolidation is relatively brief.
In parallel, biochemical investigations of the active and inactive state of CREB over time after training in different types of tasks helped determine the temporal dynamics of CREB activation underlying memory formation. Using phosphorylation of Ser-133 (pCREB) as a marker of CREB activation (175), the profile of activated CREB was demonstrated in fear-conditioning memories, including inhibitory avoidance (IA) and contextual fear conditioning (CFC). These memories are generally evoked by presenting animals to a context or cue in association with a foot shock. Memory retention is shown by measuring the generated avoidance of the context or the freezing response (134, 236). It was found that in IA a learning-specific pCREB increase in the hippocampus follows a biphasic profile (28). The first peak of activation in CA1 hippocampal neurons begins immediately after training and returns to baseline at 30 min, whereas the second peak arises between 3 and 6 h post-training and returns to baseline by 9 hr after training. This profile of increased pCREB coincided with hippocampal PKA activity in rats trained in the same task, suggesting that the CREB activation necessary for consolidation of the memory is mediated by PKA phosphorylation.
Similarly, we found a sustained pCREB increase in the hippocampus after IA training (229, 230). That increase lasted longer than the one described by Bernabeu et al. (28), perhaps because of the use of a slightly different task procedure and shock intensity. In our paradigm, the pCREB increase was evident immediately after training and was sustained at 3, 6, 9, and 20 hrs post-training. The pCREB increase seems to be due to the phosphorylation of pre-existing CREB, not to a change in the expression levels of CREB.
Stanciu et al. (224), like Bernabeu et al. (28), described a biphasic pattern in pCREB activation in the hippocampus, parietal cortex, and amygdala of mice trained in contextual fear conditioning. The first peak occurred between 0 and 30 min; the second, highest peak occurred between 3 and 6 hrs after training. Recent studies of Aplysia have also found that CREB1 levels are elevated for several hours after the induction of long-term synaptic facilitation. Moreover, CREB1 was found to bind to the promoter of its own gene, indicating a potential positive feedback in the sustained regulation of this gene (142). One possibility is that feedback loops modulate the formation of long-term memory. For example, in a positive feedback loop, phosphorylated CREB1 might induce its own gene via CREs, whereas in a negative feedback CREB2 might repress its own gene. CREB2 up-regulation via CREB1 may limit the activity of the CREB1-mediated positive feedback loop (172). Regulatory immediate early genes, once induced, may regulate the expression of upstream regulators, resulting in sustained regulation of gene expression (212).
Interestingly, it has been found that in-vivo LTP is accompanied by biphasic activation of CREB, as shown in studies of nondecremental L-LTP (216). Tetanic stimulation appears to elicit a first peak of CREB phosphorylation 30 min later. A second, more sustained peak starts about 2 hrs after stimulation and is maintained for at least 24 hrs. Similarly, in hippocampal slices subjected to HFS, the delayed onset of pCREB increases in two peaks, one at 45 min after treatment and the second at 6 hrs, which returns to baseline level at 8 hrs after induction (5). Blocking protein synthesis with anisomycin blocks only L-LTP pCREB enhancement between 2 and 8 hrs. This indicates that the sustained phosphorylation of CREB probably is mediated by one or more kinases that are activated during both an early and a late phase, with only the late phase requiring protein-synthesis-dependent phosphorylation events. Studies of rat organotypic hippocampal slices also revealed that the induction of LTP in CA1 is accompanied by a local increase in pCREB (136) is sustained for at least 4 hrs, again supporting the hypothesis that CREB is important in the late phase of LTP.
Despite all this evidence that CREB phosphorylation in Ser133 correlates with learning and memory, it is important to remember that phosphorylation of Ser 133 is a prerequisite for CREB activation, but is not sufficient to mediate CREB-dependent gene expression. Recent findings show that the transcriptional activity of CREBS can also be dramatically activated independently from Ser133 phosphorylation by the action of factors known as transducers of regulated CREB activity (TORCs) (31). As is true of the function of TORC2 in the pancreas as a coincidence detector, in neurons the recruitment of TORC1 seems to be important in the CRE-regulated transcription underlying the modulation of synaptic strength. TORC1, which is expressed in the adult brain, translocates to the nucleus upon the concomitant activation of calcium and cAMP signaling pathways and produces synergistic activation of CREB-mediated transcription. TORC1 has been found to be critically involved in the maintenance of L-LTP, suggesting that TORC1-mediated CREB regulation, like ser 133-dependent CREB activation, is a critical molecular step underlying synaptic plasticity and long-term memory (127). The function of TORC1 in long-term plasticity and in encoding and storing associative memories is intriguing because it may provide a molecular explanation of the fact that these memories require the coincidence of different input signals and translation of these inputs into changes in the number, structure, or function of synapses (127).
Along the same line of thinking, a recent study has revealed that changes in CREB function, probably resulting from coincidental integration of inputs, influence the probability that individual neurons will be recruited in encoding memory. Han et al. (87) have provided evidence that only about one-quarter of neurons of the lateral amygdala that receive sensory inputs show CREB Ser 133 phosphorylation after auditory fear conditioning, indicating that CREB activation is critical for determining which neurons are recruited into the fear memory trace. The restricted expression modulation of CREB function in a similar portion of LA neurons via viral vectors has shown that there is neuronal selection during memory formation. In other words, memory formation seems to recruit eligible neurons as a function of their relative CREB activity at the time of learning (87). Thus, following learning, CREB function appears to be critically recruited in a subset of neurons that are necessary for mediating the consolidation of learned information.
IIB. CCAAT Enhancer Binding Proteins (C/EBP), a Regulatory CREB-Downstream Immediate Early Gene Required for Synaptic Plasticity and Memory Formation
In mammals, the C/EBP family of transcription factors includes six isoforms defined by distinct genes: C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ, C/EBPε, and C/EBPζ. The C/EBP family, like CREB, Gcn4, c-Jun and c-Fos, belongs to the superfamily of “b-zip” transcription factors. This motif is positioned at the C-terminus, whereas the N-terminal generally includes the activation domain.
Gene expression regulation via C/EBPs, like that of most transcription factors, is complex and controlled at multiple levels. Because the b-zip domain is highly conserved, different C/EBPs are able to form homodimers and heterodimers. Heterodimer formation enhances the number of target sequences to which C/EBP can bind in different promoters (73, 133, 246, 253). Moreover, the trans-activation potentials of the various members differ, so that heterodimerization can produce various complexes that regulate the expression of target genes by different mechanisms. As in most families of transcription factors, not all C/EBP isoforms are activators of gene expression; indeed, some of them, such as C/EBPζ, can act as dominant negative inhibitors. Also, C/EBP complexes can regulate target gene expression by switching their function from activator to inhibitor. For example, C/EBP can heterodimerize with other members of the C/EBP family, members of different b-zip families, or members of other types of transcription factor families, including CREB, Fos, NF-kB, Gcn4, and Myc.
With the exception of C/EBPζ, the C/EBPs interact with the dyad symmetrical repeat RTTGCGYAAY, where R represents A or G and Y represents C or T. However, the C/EBP binding site may also include several variations, indicating that C/EBPs are pleiotropic DNA binding factors. C/EBPζ is an exception because two proline residues in its basic regions disrupt the α-helical structure and its ability to dimerize with other family members, resulting in a heterodimer that cannot bind to the C/EBP binding site in the promoter of target genes (208). These heterodimers recognize different consensus sequences in the promoters of a subset of genes activated in stress conditions (242). Thus, C/EBPζ can function either as a dominant negative inhibitor of C/EBP function or as a direct activator of stress genes.
The gene structure of C/EBPs is simple: C/EBP α, β, γ, and δ are intronless, while C/EBPε and ζ contain two and four exons, respectively (6, 41, 201, 253). Each of the six C/EBP genes can generate the expression of several proteins by alternative use of translation initiation codons in the same mRNA molecule, alternative use of promoters, and differential splicing. (201) (Fig. 4). In addition, post-translational modifications of C/EBPs are important in directing their function and stability.
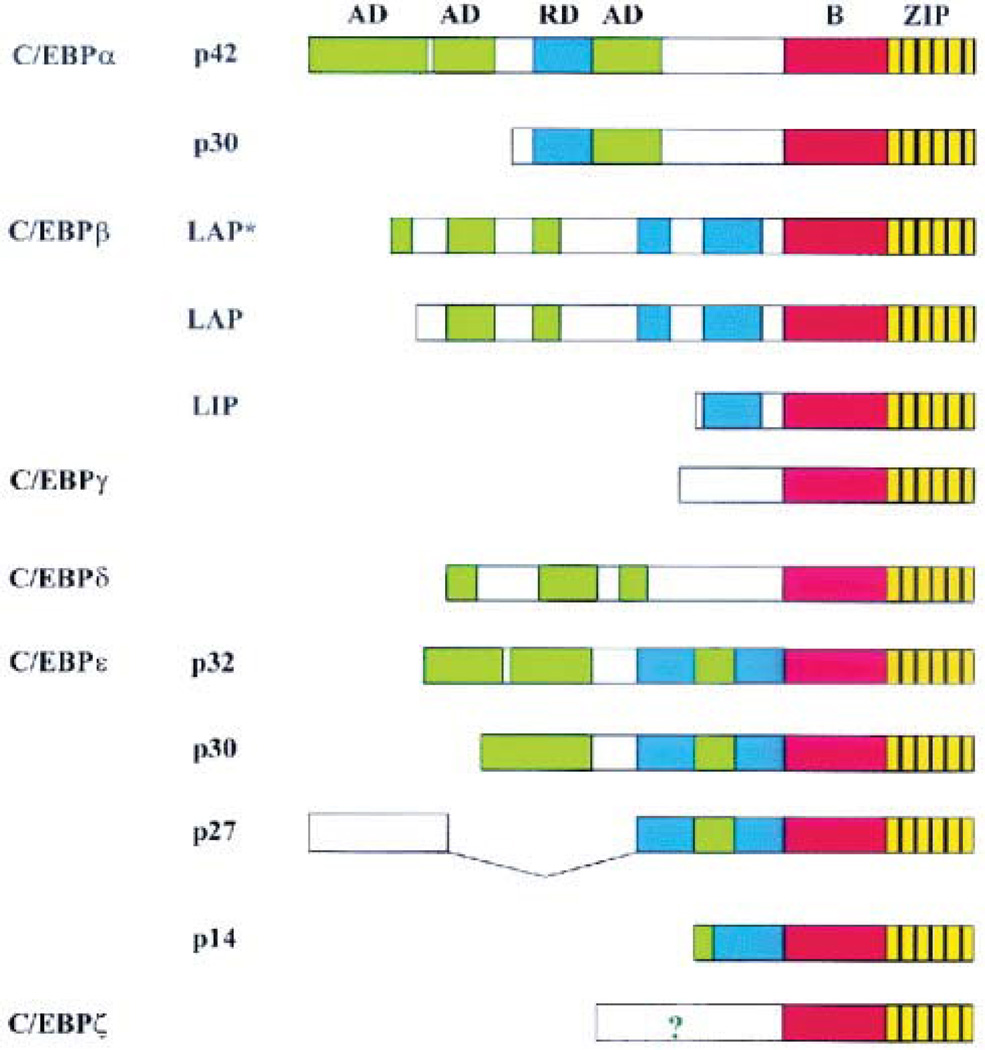
Figure 4. Genomic organization of the mammalian C/EBP family members.
The leucine zipper (ZIP) is shown in yellow, with black vertical lines indicating the leucine residues, and the basic region is colored red. The position of the activation domains (AD) and negative regulatory domains (RD) are shown in green and blue respectively. ? indicates that the N-terminus of C/EBPζ contains an activation domain, although its exact position remains to be determined. [From Ramji and Foka (201)].
C/EBPs are expressed in various tissues, including CNS, adipose, liver, intestine, lung, adrenal gland, peripheral blood mononuclear cells, and placenta. They have key functions in differentiation, the inflammatory response, liver regeneration, metabolism, synaptic plasticity, and memory, as well as in a variety of cellular functions, including stress, growth, and apoptosis. The regulatory and functional roles of C/EBPs in various tissues have been reviewed previously (131, 154, 201, 210, 214). My focus here is on current understanding of the expression regulation and functions of C/EBP family members in synaptic plasticity and learning and memory.
The first evidence that C/EBPs are expressed in neurons and involved in long-term synaptic plasticity underlying memory formation came from studies of the invertebrate Aplysia californica. As noted earlier, Aplysia co-culture systems are used in in-vitro modeling of the short- and long-term synaptic responses that occur during simple forms of memory. Experiments in co-culture systems were instrumental in determining the fundamental role of C/EBP in long-term synaptic plasticity. Indeed, this in-vitro system pioneered the identification and characterization of several molecules and molecular mechanisms underlying memory formation (10, 21, 117).
Two splicing isoforms of the same intronless C/EBP gene were cloned from the Aplysia CNS (ApC/EBP) (9). Biochemical characterization showed that these transcripts, as compared to the mammalian C/EBP isoforms, were more homologous to C/EBPβ. This was indicated by conserved phosphorylation consensus sequences for MAPK and calcium calmodulin Kinase II (CamKII), and the homology of the b-zip domain (9). Notably, expression levels of ApC/EBP were nearly undetectable in resting conditions both in vivo and in vitro. However, ApC/EBP mRNA was rapidly transcribed and translated upon activation of the cAMP pathway or application of 5HT. The regulation of this induction is typical of an immediate early transcript, suggesting that 5HT or cAMP-mediated ApC/EBP expression is part of a cascade of gene expression. Using an approach similar to that of Dash et al. (52), Alberini et al. (9) showed that inhibition of ApC/EBP expression or function by means of antisense sequences, blocking antibodies, or C/EBP decoy results in disruption of long-term facilitation, while short-term facilitation is left intact. The critical function of ApC/EBP during long-term facilitation in cell culture lasts more than 9 hrs, indicating that the ApC/EBP-dependent transcriptional phase essential for long-term facilitation is long-lasting.
Subsequent studies by Lee et al. (135) confirmed these findings, showing that blocking the expression of ApC/EBP by RNA interference (RNAi) disrupts long-term facilitation without affecting short-term facilitation. These authors also showed that a single pulse of serotonin on sensory neurons that over-express ApC/EBP results in long-term facilitation instead of short-term facilitation, indicating that the expression of ApC/EBP in cultured sensory neurons is sufficient for mediating the consolidation of short-term into long-lasting facilitation (135). Together, these findings show that a cascade of gene expression in which transcription factors regulate the expression of other transcription factors, which in turn regulate the expression of effector genes, is an essential molecular signature for long-term memory formation (9, 77). This important understanding revealed that the gene expression underlying memory formation is regulated by processes that are mechanistically similar to those underlying cell differentiation and development, in which changes in phenotypic expression are due to changes in epigenetic mechanisms. Recent work has confirmed that changes in chromatin structure and posttranslational modifications accompany long-term memory formation (79, 138).
Numerous studies done in parallel revealed that, like ApC/EBP, CREBs expressed in Aplysia are capable of transforming short- into long-term facilitation, suggesting that activation of this pathway is sufficient to stabilize synaptic plasticity underlying long-term memory formation (25). Later studies in mammals demonstrated that C/EBP isoforms, particularly C/EBPβ and C/EBPδ, have an evolutionarily conserved and critical function during memory formation (46, 225, 230, 231). Rats trained with inhibitory avoidance, during which, as described earlier, the rats learn to avoid a context previously associated with a foot shock, show enhanced expression of pCREB, followed by an increase in both C/EBPβ and C/EBPδ in the same neuronal populations of hippocampal CA1 and dentate gyrus regions (230). pCREB increases immediately after training; this enhancement is sustained for at least 20 hrs. C/EBPβ and C/EBPδ expression significantly increase between 6 and 9 hrs after training. These increases are sustained for at least 28 hrs, returning to control levels by 48 hrs after training. The learning-dependent increases in pCREB, C/EBPβ, and C/EBPδ are likely to be mediated by modulatory neurotransmitters, which requires an intact fornix (229), but the nature of the modulations involved are still unclear.
C/EBPβ has an essential function in the hippocampus during the consolidation phase of inhibitory avoidance, since knocking down hippocampal C/EBPβ expression via stereotactic injections of antisense oligodeoxynucleotides (ODNs) disrupts memory retention. In agreement with the change in the expression profile of C/EBPβ, the functional requirement for hippocampal C/EBPβ during IA memory consolidation is transient, but lasts more than a day. This indicates that the transcription phase regulated by C/EBPβ, which is required for memory formation, is active for a relatively long time (231). As in Aplysia cultures, C/EBPβ expression in the hippocampus occurs after CREB phosphorylation (20).
Another C/EBP, C/EBPδ isoform, is also critical in memory formation. Sterneck et al. (225) have shown that C/EBPδ knock-out mice show selective enhancement of contextual fear conditioning but normal water-maze memory. These results imply that distinct C/EBP isoforms exert different functional roles in memory formation and that C/EBPδ can act as an inhibitor of the gene expression required for memory formation. Similar conclusions have been obtained in another study, which found enhancement of memory and LTP with expression in the forebrain of a broad dominant-negative C/EBP (EGFP-AZIP). However, this dominant negative, like C/EBPs, can also interact with inhibiting isoforms of ATF4, a distant member of the C/EBP family of transcription factors. Hence, the possibility that other factors may involved in this outcome cannot be excluded (46). Together, these studies suggest that the relief of C/EBP or C/EBP-like-mediated inhibitory mechanisms lowers the threshold for hippocampal-dependent LTP and memory storage in mice. However, it should be kept in mind that because of the spatial and temporal limitations of the knock-out approaches and the nonspecific targeting of general C/EBP inhibitors, it is also possible that the enhancement of memory is a result of compensatory effects or the targeting of other C/EBP inhibitor isoforms.
How is the expression of C/EBPs regulated in neurons? Studies indicate that during long-term synaptic plasticity and memory formation, C/EBPs critically recruit both cAMP and Ca++-dependent signal transduction pathways (266). The first example of this regulation was observed in the pheochromocytoma PC12 cell line, where C/EBPβ has been found to promote neuronal differentiation. In these cells, C/EBPβ is expressed in the cytoplasm and translocates to the nucleus upon cAMP stimulation (164). Although the contribution of the cytoplasm to nucleus translocation of C/EBPs has not yet been investigated, interestingly, in both Aplysia and hippocampal neurons, C/EBPs are localized in the cytoplasm, suggesting that nuclear translocation may be a mechanism that functionally links the distal (synaptic) compartments to the nuclear district, resulting in gene-expression regulation.
Another level of regulation of the C/EBP function in neurons occurs through the activity of kinases, including MAPK, CaMK II and IV, and protein kinase C (259). These kinase-dependent regulations participate in the activation of C/EBPs as transcription factors, as well as in their degradation. For example, ApC/EBP is degraded through the ubiquitin-proteasome pathway, but its phosphorylation via MAPK prevents its proteolysis (259). Thus, it seems that phosphorylation of ApC/EBP by MAPK synergistically acts on two levels to ensure that C/EBP-dependent gene expression is sufficiently prolonged during the consolidation phase.
Which target genes do C/EBPs regulate during memory formation? This question is both important and technically difficult to address. As with all transcription factors, it is the convergence of different signaling pathways that acts in concert in space and time to dictate the pattern of the expressed target genes. Thus, it is important to be technically able to identify the combinations of transcription factors that, over time, are involved in regulating a given function. With this knowledge, we should be able to determine why C/EBPs, first discovered in the induction of the acute-phase response in the liver, are also fundamental for long-term memory formation. In summary, as pointed out earlier, C/EBP and CREB family members participate in a fundamental, evolutionarily conserved cascade of events that are required for the consolidation of new memories (11, 42) (Fig. 5).
Figure 5. Schematic representation of the CREB-C/EBP pathway activated during memory formation.
Stress, neurotransmitter release, growth factors and membrane depolarization are among the stimuli that activate intracellular signal transduction pathways that can lead to the activation of the CREB-dependent cascade. An important step of this activation is the phosphorylation of CREB (pCREB), particularly in its Ser133 residue. The functional activation of CREB leads to the expression of target genes, among which there are immediate early genes (IEGs), such as the transcription factor C/EBP, which, in turn, presumably regulates the expression of late response genes [From Alberini et al. (13)].
IIC. Activating Protein 1 (AP-1), a Marker of Neuronal Activation
The transcription factor AP-1 is composed of b-zip dimers of members of the Jun, Fos, and ATF families. Fos proteins include Fos, Fra1, Fra2, and FosB; members of the Jun family include Jun, JunB, and JunD. Fos proteins do not form homodimers but can heterodimerize with members of the Jun family. However, Jun proteins can both homodimerize and heterodimerize with other Jun or Fos members to form functionally active complexes. Jun proteins also heterodimerize with other transcription factors such as members of the ATF family (83) and other basic zipper-containing transcription factors such as CBP, MyoD, Nfat, and c-rel (95).
AP-1 is involved in both basal and inducible transcription of several genes that bear the AP-1 consensus site 5’-TGAG/CTCA3’, also known as TPA-responsive element, or TRE, in their promoter region. AP-1 can also bind, but with lower affinity, to the CRE element. As with other transcription factors, variants of the canonical sequences can also be recognized and contribute to the differential function of AP-1 at various regulatory sequences (Fig. 6).
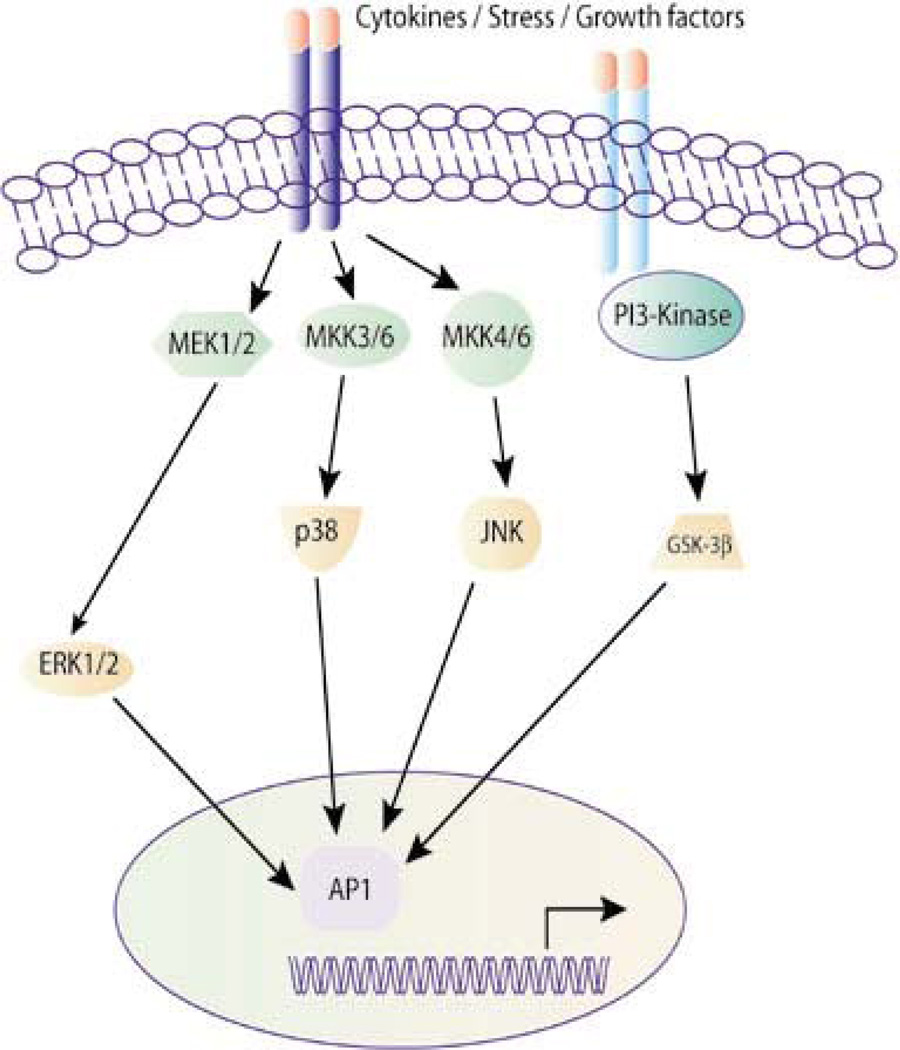
Figure 6. Schematic representation of AP-1 activation pathways.
A multitude of stimuli and environmental insults including cytokines, stress and growth factors can regulate a considerable number of intracellular processes that critically involve AP-1. The appropriate composition of subunits in the AP-1 dimer is determined by the nature of the extracellular stimulus and by the MAPK signaling pathway that is consequently activated [From Panomics © 2007 (189)].
AP-1 mediates numerous types of responses to a variety of physiological and pathological stimuli, including cytokines, growth factors, stress signals, bacterial infections, viral infections, and oncoproteins. Gene knockout experiments have shown that Fos and Jun proteins mostly have overlapping functions, but some isoforms have also been found to have unique functions that cannot be compensated for by other family members. The functions of Fos and Jun family members are multiple and depend on the specific cell type in which they are expressed. For example, in the nervous system the upregulation of c-Jun and related transcription factors is a common event that not only underlies development and adult brain functions, but also occurs during injury and repair (95, 200). Functions that critically involve AP-1 in the CNS are neuronal differentiation and survival during normal embryonic and postnatal development. AP-1 in the CNS is also involved in transplantation, seizures, addiction, pain, cerebral ischemia and stroke, axotomy and other forms of trauma, and posttraumatic repair (45, 58, 64, 74, 75, 89, 92, 93, 94, 106, 121, 124, 161, 178, 181, 199, 249, 255). Ap-1 has also been implicated in some cases of LTP and/or LTD, as well as memory formation (3, 237). Several excellent reviews on the different members of the AP-1 family are available (57, 76, 95, 178, 183).
C-Jun expression is strongly increased in seizures and in some but not all cases of LTP and/or LTD. In fact, the function of jun in LTP is evident only with specific technical protocols. C-fos mRNA has also been found to be upregulated in response to a variety of neuronal activation protocols, including LTP. However, because a multitude of stimulations, including sensory, electrical, and epileptogenic, are associated with an increase in c-fos, it appears that c-fos expression correlates with neuronal activity rather than synaptic plasticity. Often, it is difficult to differentiate these two processes. During generalized acute seizures, increases in mRNA encoding for junB, c-fos, and krox-24 are found in the dentate gyrus within 10–15 min (130, 264), subsequently in the hippocampal CA1 and CA3 areas, and, finally in the cortex (49, 251). Expression of the corresponding proteins is considerably slower. However, an injury response may also contribute to the temporal dissociation between mRNA and protein expression. The mRNA levels decline to basal concentrations within 1–2 hrs in the dentate gyrus and 2–4 hrs in the CA1/3 areas. Compared with the other AP-1 induced transcription factors, the up-regulation of c-jun is more moderate and prolonged, persisting for 24–72 hrs (49, 115, 221). On one hand, chronic electroconvulsive seizure is reported to correlate with decreased expression of Fos and Jun (39, 254); on the other hand, constitutively epileptic animals have higher cortical and hippocampal AP1 transcription factor levels than do their nonepileptogenic parents (263).
Both correlative and functional studies have provided evidence of the function of AP-1 in the formation of several types of long-term memories. However, conflicting results have been reported. Increased expression of c-Jun in the adult brain has been reported during novel experiences, short- and long-term memory formation, and both painful and rewarding experiences (3, 92, 190, 255). Passive avoidance in newly hatched chicks results in a four-fold increase in c-jun mRNA and protein levels in the forebrain, particularly the lateral intermediate ventral hyperstriatum (19) and lateral paraolfactory nucleus (63). In mice, training in an appetitive bar-pressing task induces a spatially selective expression of c-Jun along with c-Fos in the hippocampal CA1, CA3, and dentate gyrus regions (97, 98), whereas continuous training with trace eyeblink conditioning for 24 hrs or longer can induce a down-regulation in c-Jun levels (56). Antisense ODN-mediated knock-down of c-jun, but not junB, inhibits sequence-specific learning following an aversive stimulus using the foot shock-motivated brightness discrimination (237). Conversely, complete deletion of the c-jun gene by neural excision does not interfere with spatial learning in a water maze or in fear conditioning paradigms (199).
Contrasting results have been found in the expression of c-fos with spatial tasks. However, in other types of tasks, the function of c-fos appears to be critical. For example, acquisition of socially transmitted food preference and appetitively motivated spatial learning correlates with an increase in c-Fos as well as pCREB in the hippocampus. Also, c-Fos expression is necessary for the formation of these memories (50, 220). Moreover, c-fos has a critical function in fear-induced immobility, morphine-induced conditioned place preference, conditioned taste aversion, and passive avoidance (128, 168, 179). It is consistent with these findings that a correlative increase in c-Fos expression occurs after learning. For example, Fos is present in the dorsal striatum of rats trained on a variable position reward task in the radial arm maze and in the hippocampus, entorhinal cortex, and primary visual cortex 30 min after spatial learning (17, 81, 90).
The temporal profiles of c-Fos expression in different neuronal populations have been investigated using several memory tasks. It appears that c-Fos induction occurs after early training, within minutes and, that, like the transcription factor Zif268, c-Fos becomes most evident during the first few trials, decaying after repeated trials. For example, a comparison of sham-trained mice with mice trained in an appetitive operant conditioning task had increased c-Fos levels in CA3, the anterior cingulate, and the occipital and parietal cortex at 60 min, but not 120 or 180 min post-training on day 1. On day 2, c-Fos was increased in CA1, CA3, the dentate gyrus, entorhinal cortex, posterior cingulate cortex, and striatum 60 min after training. No differences were found at the later times. On day 5, no differences were detected between trained and control groups (29).
Studies with hippocampal LTP have reported only a partial correlation between LTP and c-Jun expression, as was found in behavioral experiments. In fact, correlative changes in c-jun mRNA or protein and LTP have been detected in some (49, 257) but not all studies (255). There is also evidence supporting the function of c-Jun in synaptic plasticity in the larval neuromuscular junction of Drosophila (212). In this model system, inhibition of c-Jun function by overexpression of a dominant negative isoform inhibits synaptic plasticity and reduces both synaptic strength and synaptic bouton number via CREB, activity dependent factor 1 (ADF1), and fasciclin 2 (FasII) mechanisms. Thus, AP-1 family members seem to be critically recruited during the activation of neuronal circuits leading to long-term changes such as those underlying long-term memory formation.
IID. Zif 268, a Marker of Neuronal Plasticity?
The transcription factor zif268 belongs to the Egr family; it is also known as Krox24, NGF-I-A, Egr-1, TZs8, and Zenk. It was first discovered as an immediate early gene regulated by nerve growth factor and in response to serum treatment (Fig. 7). The zif268 gene encodes for two polypeptides of 82 and 88 kDa. These polypeptides have different translational starting sites and belong to the structural family known as zinc fingers. This structural family is characterized by a domain that folds into three zinc finger structures that mediate DNA binding to the element 5’-GCGC/GGGGCG-3’. The high homology of the finger sequence among all members of the family implies that they may bind to the same sites of at least a subset of the same target genes.
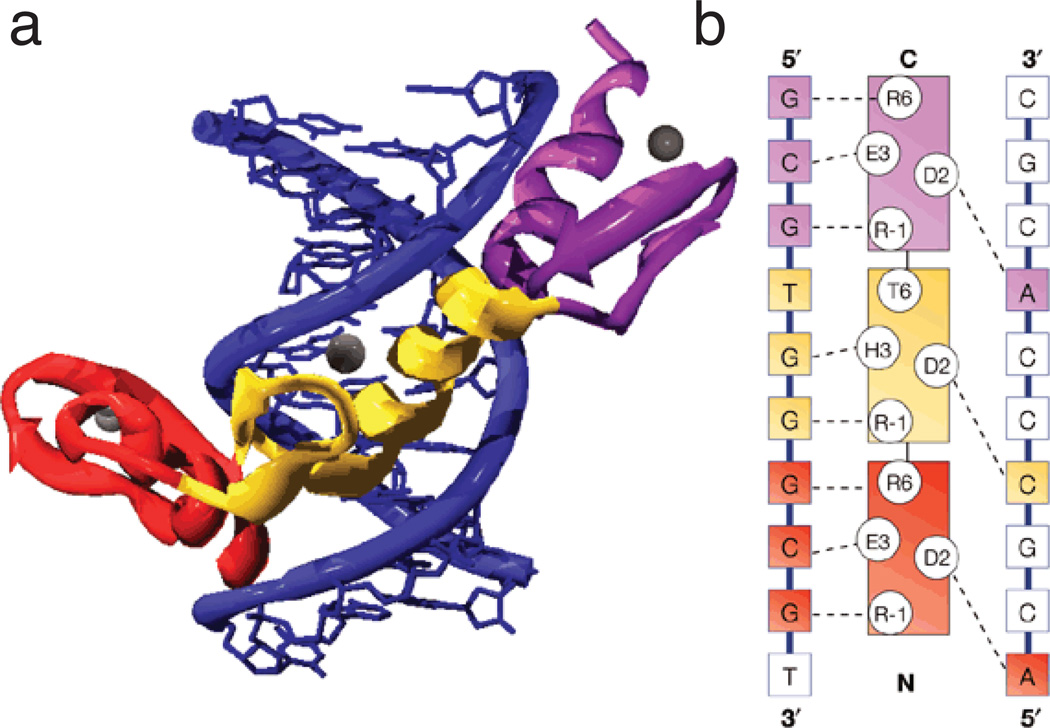
Figure 7. Modular interactions between zinc fingers and DNA.
a) The Zif268–DNA complex showing the three zinc fingers bound in the major groove of DNA. The DNA is blue and fingers 1, 2, and 3 are red, yellow, and violet respectively. Zinc ions are shown as grey spheres. (b) A diagram showing the sequence-specific protein–DNA interactions between Zif268 and its DNA-binding site. The recognition helices of the three fingers are represented in the centre of the panel and the bases on the two strands of the DNA site are shown on either side. The identity of key residues on the recognition helices (positions -1, 2, 3 and 6 with respect to the start of the helix) are also shown using the single-letter code. Contacts observed in the crystal structure are represented as dashed lines. The fingers and bases that they contact are color-coded using the same scheme as in (a). The fingers are spaced at three-base-pair intervals and tend to contact three adjacent bases on one strand of DNA and one base on the other strand. [From Jamieson et al. (103)].
In the rat brain, Zif268 is constitutively expressed in the neocortex, primary olfactory cortex, entorhinal cortex, amygdaloid nuclei, nucleus accumbens, striatum, cerebellar cortex, and hippocampus. In the hippocampus, expression of zif268 is high in the CA1 region, less in CA2-CA3, and minimal in the dentate gyrus where, however, it can be induced in response to a various stimuli, including memory, seizure, kindling, brain injury, neurodegeneration, apoptosis, stress, and drug tolerance and dependence (54, 123, 210).
Members of the Egr family show a similar pattern of basal expression. Except for Egr2, which is predominantly localized in the nucleus, the family is expressed in the nucleus and cytoplasm (146). The expression of zif268 mRNA is strongly induced during LTP at dentate gyrus synapses 30–60 min after tetanic stimulation of the perforant path. This induction requires the function of NMDA receptors (65, 150, 203). Interestingly, the function of zif268 seems to be associated with the maintenance rather than the induction of synaptic plasticity. In fact, the post-tetanic increase in zif268 correlates more strongly with the persistence of LTP than with its magnitude of induction (3, 109). It is consistent with this finding that zif268 mutant mice show a normal induction and early phase of dentate gyrus LTP, but faster decay of the LTP late phase. Thus, it was initially concluded that zif268 is? a marker of plasticity (35, 109). In addition to the hippocampus, several other brain regions have been found to display an increase in zif268 mRNA following LTP. These include, for example, the visual cortex after stimulation of the thalamocortical tract (99) and the insular cortex after stimulation of the basolateral amygdala (108).
Some controversial results have been obtained concerning the regulation of zif268 expression after long-term synaptic plasticity. For example, zif268 does not seem to change in the CA1 region of the hippocampus when LTP is induced in vivo by high-frequency stimulation, but significantly increases after in-vivo tetanic stimulation. No change in zif268 expression was reported after LTP was induced in slices of CA1 (65). However, it is important to remember that for zif268, as for all other inducible transcripts and proteins, the general pattern of gene expression and its regulation differ in slices as compared to the intact brain (232). There also have been conflicting results with respect to the expression of zif268 not only in experience-dependent plasticity in the visual cortex (108, 149), but in kindling in the amygdala (40). In both cases, while acute presentation of the stimulus correlated with the expression of zif268, similar experiments on zif268 mutant mice demonstrated no difference as compared to wild-type littermates. However, as noted in reference to CREB and C/EBP studies, a possible explanation for the discrepant results obtained with acute approaches versus the use of general knock-out animal models is that in the latter functional compensation may overcome the deficit produced by gene deletion. Although zif268 is the most extensively characterized member of the Egr family, others, such as Egr 2, have also been found to be regulated following LTP (252, 258). The expression of Zif268 seems to be regulated in response to various behaviors, including learning and memory. Zif268 is rapidly regulated after associative learning such as song learning in birds, fear-conditioning and spatial learning in rats, and in visual-paired association in monkeys (33, 59, 81, 84, 85, 105, 147, 158, 159, 185, 209, 238, 239).
In birds, expression of zenk, the avian homologue of zif268, is regulated in regions that process not only song learning, but also song perception. Since zif268 has been generally linked to neuronal plasticity in the mammalian brain, it is believed that zenk may be linked to molecular and cellular mechanisms that underlie the experience-dependent modification of song-responsive circuits (104, 160).
In rodents, fear-conditioning training correlates with upregulation of zif268 in the CA1 region of the hippocampus and in the amygdala (147). However, because similar increases occur in matched controls, it is likely that the change in zif268 is generally associated with the experience of novelty (85). The characterization of zif268 expression in spatial learning has indicated that in multiple trial learning tasks the regulation of this transcript seems to be restricted to an initial phase. With repeated trials, its upregulation is attenuated (81, 202, 255).
Genetic studies in mice have also supported the conclusion that Zif268 is critical for memory formation. Zif268 knockout mice show impaired long-term memories but intact short-term retention. Long-term memory deficits have been found in various tasks, including spontaneous alternation, social transmission of food preference, object recognition, spatial learning, and conditioned taste aversion (108). Consistent with these findings, heterozygous mice have memory deficits that fall midway between the performance levels of knockout and wild-type mice (108). As with CREB mutant mice, when zif268 mutants are exposed to overtraining behavioral protocols, they gain the ability to learn and form memories, suggesting that mechanisms of molecular compensation or the use of different learning or memory strategies can compensate for the loss of essential genes. It has been debated whether the regulation of zif268 expression also occurs in association with the retrieval of fear-conditioning memory, a process that has been more closely scrutinized over the last few years. Significant changes in amygdala zif268 levels after the retrieval of contextual fear conditioning have been reported in some but not all cases (85, 234, 247). It, remains to be established whether the regulation of zif268 is specifically linked to associative learning after acquisition, the reactivation of memory, or both, or whether it is, instead, associated with processes that are prerequisites to learning and memory consolidation.
IIE. NFkB: From Synapse to Nucleus via Retrograde Traveling
Nuclear factor kB (NFkB), a transcription factor first characterized in lymphocytes, is a ubiquitously expressed protein. NFkB belongs to the Rel family of transcription factors, which consists of five members: p50, a product of the NFkB1 gene; p52, a product of the NFkB2 gene; p65, also known as RelA; c-Rel; and RelB. The Rel family members function as dimers can either homodimerize or heterodimerize (Fig. 8). They also share a Rel homology domain containing the following structural elements: DNA binding, dimerization, nuclear localization, and an inhibitory kB (IkB) interaction region. The NFkB typical DNA binding element is 5’-GGGACTTTCC-3’.
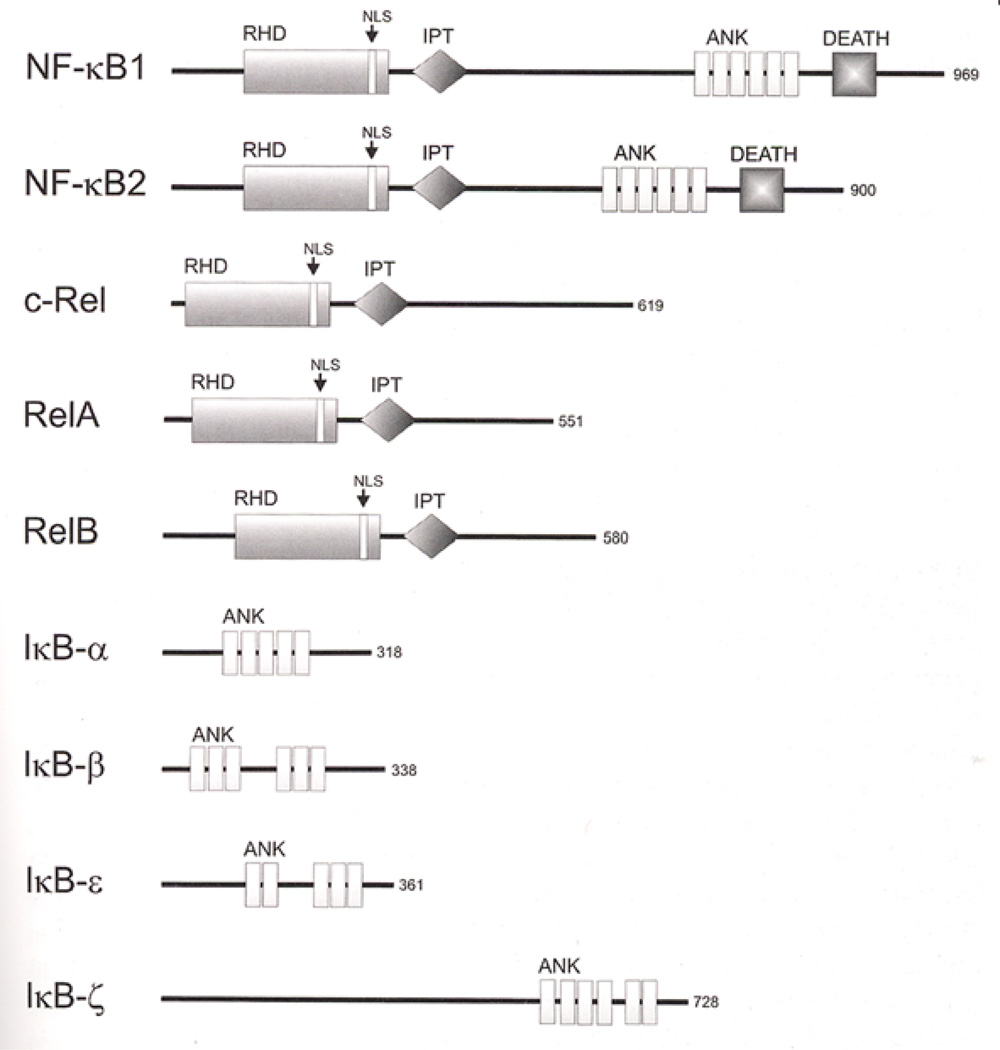
Figure 8. Structure of the NFkB subunits.
Domain motifs are indicated. Protein size is given on the right as amino acids. RHD: Rel homologous domain. NLS: nuclear localization signal; IPT: Ig-like, plexin, transcription factors; ANK: ankyrin repeats; DEATH: DEATH domain, found in proteins involved in cell death. [From Kaltschmidt et al. (113)]
Compared to most transcription factors, NFkB has an interesting feature; i.e., in resting conditions dimers are retained in an inactive form in the cytoplasm by binding to IkB inhibitory proteins, of which IkBα and IkBβ are the most abundant. IkBs act by masking nuclear localization signals (NLS) within NFkB subunits (88). Upon stimulation, the IkB kinase complex becomes activated and phosphorylates IkB, leading to its ubiquitination and subsequent degradation. This degradation allows NFkB to translocate to the nucleus and regulate the expression of its target genes.
A critical function of NFkB was first found in the immune system, where NFkB-dependent gene expression is critical during inflammation and host-defense (141). More recently, it has been demonstrated that NFkB is important in synaptic plasticity, as well as learning and memory. NFkB is expressed in neurons and nonneuronal cells such as glia and Schwann cells. Indeed, expression of multiple NFkB family members has been found in different types of cells in the CNS. The subunits most commonly expressed in neurons are p50-p65 heterodimers and p50 homodimers, although only p65/p50 heterodimers have been found in synaptosomal preparations (155, 157). Thus far, only a relatively small number of NFkB target genes have been found in neurons; these include Ca2+-calmodulin-dependent protein kinase II (CaMKII) δ, brain-derived neurotrophic factor, μ-opioid receptors, neural cell adhesion molecule, inducible nitric oxide synthetase, and amyloid precursor protein (186, 207).
NFkB was initially reported to be constitutively active in the hippocampus, cerebral cortex, and cerebellar neurons. However, evidence suggested that NFkB is activated upon excitatory synaptic transmission through the Ca2+-responsive signaling cascade and the recruitment of CaMKII. Interestingly, NFkB also becomes active in response to dopamine. The latent form of NFkB can become active in response to stimuli that include glutamate, kainate, amyloid β peptide, tumor necrosis factor α, brain injury, oxidative stress, and depolarization.
The function of NFkB in LTP has been established by several lines of evidence. LTP induced by HFS or low-frequency stimulation (LFS) of hippocampal slices correlates with an increase in the mRNA of both p50 and p65 and a decrease of IkB mRNA (155); pretreatment with NFkB DNA element decoy interferes with the induction of both LTP and LTD (8, 261). The in-vivo-induced perforant path LTP in the mouse hippocampus is accompanied by enhancement of the NFkB binding to its DNA binding elements in the hippocampal nuclear extract (68). An incremental increase in NFkB active form occurs in granular and pyramidal cells in the hippocampus after LFS and HFS-LTP, respectively (155). Similar investigations of mice with genetic disruption of NFkB restricted to the forebrain confirm that this transcription factor is critical for synaptic transmission, L-LTP induced by theta burst stimulation, and induction of LTD (114).
In-vivo experiments on different species have shown that NFkB is critically important in several types of long-term memories. In the crab _Chasmagnathus_habituation of the escape response elicited by a fear stimulus (passage of an opaque figure representing a potential predator) correlates with an increase in the activated form of an NFkB homolog in brain nuclei and isolated synapses (66, 67). Also, injection of an IkB kinase complex inhibitor blocks NFkB activation and disrupts the escape memory, indicating that NFkB is essential for this memory (162). Further studies provided evidence that NFkB is also recruited for reconsolidation of the crab escape memory since, blocking NFkB after memory recall results in amnesia (163).
Similar results have been obtained with mammalian memory systems. Mice lacking the p65 subunit of NFkB and tumor necrosis factor receptor 1, which is necessary for survival of the p65 knockout, have impaired spatial radial maze memory (156). However, as with other transcription factors described earlier, this impairment is overcome if the animals undergo overtraining protocols. Again, as with other transcription factors, the function of NFkB is not found in all memories. Some types of memories, but not others, appear to be readily disrupted by NFkB inactivation. Thus, whereas NFkB knockout mice have impaired radial maze memory, c-Rel knock-out mice have normal fear conditioning memory; intact memory is also generally found in mice lacking the p50 NFkB subunit (119, 120, 137). On the other hand, intra-amygdala administration of NFkB decoy disrupts the fear-potentiated startle response (261). Inhibition of NFkB with decoy elements or the injection of pharmacological inhibitors into the hippocampus disrupts IA memory (69). Forebrain inhibition of NFkB function obtained in mice by overexpression of IkB affects spatial memory as assessed by water maze. In these mice, both the mRNA of the catalytic subunit of PKA and CREB phosphorylation are significantly decreased, suggesting that NFkB affects activation of the CREB pathway during memory consolidation (114).
Further evidence has confirmed that NFkB has a critical function in memory reconsolidation in mammals. The use of injections of NFkB decoy to achieve intracerebral or intrahippocampal inhibition of either the kinase that activates NFkB or NFkB binding to the endogenous promoter disrupts the reconsolidation of IA memory (32). Taken together, the evidence regarding NFkB strongly argues its function in synaptic transmission, regulated synaptic plasticity, and memory formation.
One intriguing aspect of NFkB expression in neurons that clearly merits elucidation is its localization not only in the cytoplasm, but also at the synaptic compartment where, as indicated earlier, it appears to be mainly in the form of p50-p65 heterodimers. It will be important to determine whether this localization serves the function of retrograde communication to the nucleus or reflects a novel nontranscriptional function. Evidence indirectly supports the former hypothesis: a p65-GFP fusion protein expressed in hippocampal neurons migrates from distal processes to the nucleus following glutamate or kainate stimulation (248); also, NFkB translocates from distant processes to the nucleus by retrograde movement (156) that is likely to be mediated by dynei or dynactin motor complex (167).
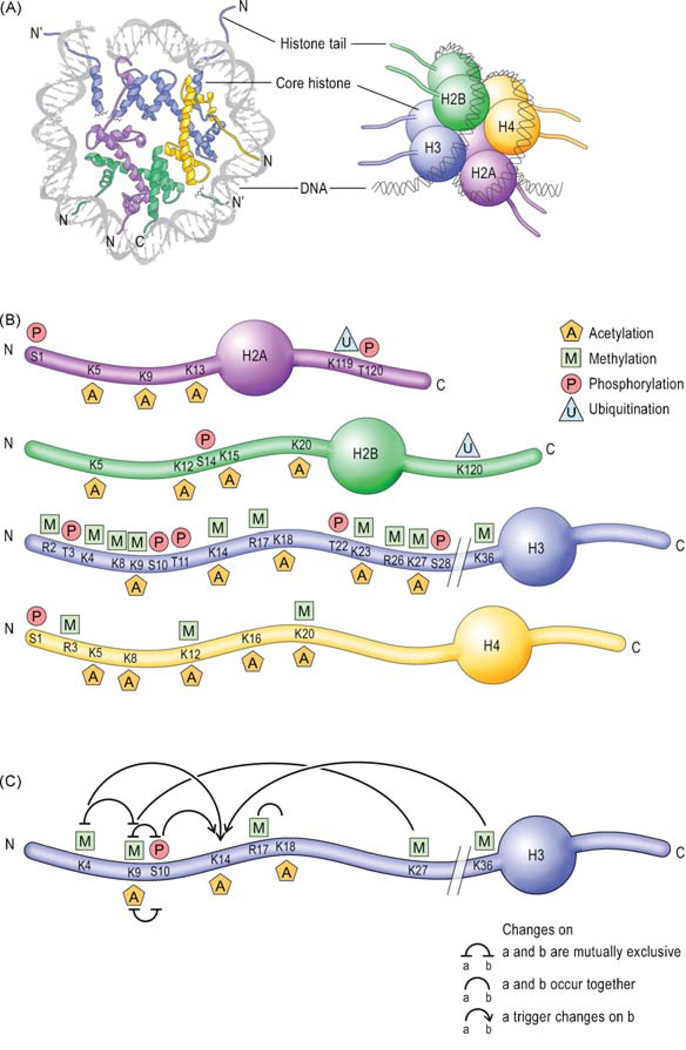
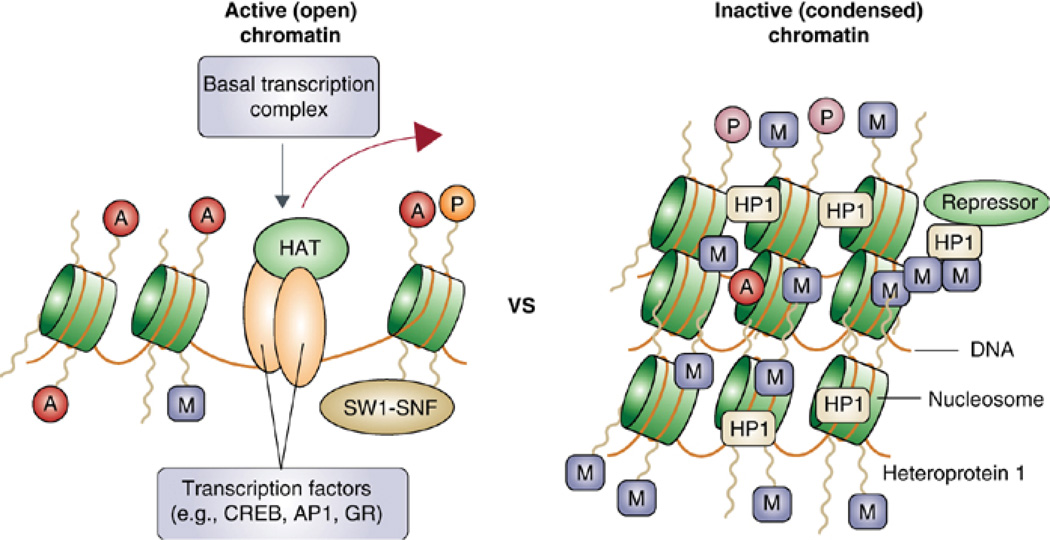
III. Transcription Regulation: Chromatin Remodeling in Synaptic Plasticity and Memory
Transcriptional cascades, whereby regulatory immediate early genes are critically recruited (9, 77, 231), are required for long-term synaptic plasticity and long-term memory formation. Thus, also chromatin changes must be critically involved. Recent studies have focused on the chromatin changes that are involved in long-term modifications underlying memory formation. Several studies have demonstrated that chromatin remodeling is critically involved in learning and memory, as well as long-term neuronal responses underlying psychiatric conditions such as drug addiction, stress, epilepsy, and depression (1, 152, 228).
Chromatin is the complex of DNA and histones that, in eukaryotic cells, constitutes chromosomal DNA; the fundamental packing unit is the nucleosome. As noted earlier, the initial step in gene-transcription activation involves recognition of the promoter and enhancer region by sequence-specific DNA binding factors. These factors can also recruit chromatin remodeling factors to the template. Chromatin-remodeling factors catalyze the mobilization of nucleosomes, which is needed for the binding of additional transcription factors and co-regulators to the DNA template. In addition, the promoter- and enhancer-binding factors mediate, directly or indirectly, the association of acetyltransferases (e.g., CBP/p300) that modify core histones and other DNA-associated proteins essential for transcription initiation. Histone acetylation likely acts through multiple mechanisms to promote the unfolding of chromatin, modulate the affinity of DNA-binding factors, and regulate activities of transcription factors and cofactors (Figs. 9 and 10). In turn, the DNA transcription-factor complex interacts with RNA Pol II to facilitate communication with the basal transcriptional machinery. Direct interactions also occur between the TAF subunits (TBP-associated factors) of the TFIID complex and promoter-enhancer-binding factors. Many, and possibly all, of these interactions are required to achieve productive transcription initiation.
Figure 9. Epigenetic marks on histone tails and DNA.
A, left) View of the nucleosome down the DNA superhelix axis showing one half of the nucleosome structure. (Right) Schematic representation of the four-nucleosome core histones, H2A, H2B, H3 and H4. (B) Schematic representation of the N- and C-termini of the core histones and their residue-specific epigenetic modifications. (C) Crosstalk between epigenetic modifications on the H3 N-terminus tail. The relationship between the different residues is based on recent literature. C, C-terminus; N, N-terminus. [From Gräff and Mansuy (82)].
Figure 10. Differential states of chromatin.
Chromatin can either be open (i.e. active, allowing gene expression) or condensed (i.e. inactive, repressing gene expression). This change in state is mediated by the modifications to core histone proteins. Histone acetylation (A) is associated with chromatin relaxation and the binding of transcription factors and co-activators, such as HATs (histone acetyl transferases) and SWI-SNF proteins that mediate the movement of nucleosomes along a strand of DNA. Histone methylation (M) results in condensed chromatin and transcriptional repression. [From McClung and Nestler (152)].
It is now clear that even the basal transcription factors and the chromatin template participate in gene-selective transcription and that complexes initially identified in general nucleosome assembly are recruited for gene-specific chromatin remodeling and gene expression (240). The mechanisms that regulate transcription are complex. Factors can alternatively act as activators or repressors, depending on their context or interacting partners. Regulatory complexes having apparently distinct functions in transcription may be linked by sharing common subunits. In addition, reversible chemical modifications of chromatin, such as methylation of DNA or histones, acetylation, or ubiquitination can variably affect gene expression. It seems likely that these different covalent modifications of regulatory proteins act in combination to provide multiple dimensions of transcription control.
Recent evidence indicates that both histone acetylation and phosphorylation are critically involved in LTP as well as memory formation. Histone acetylation is a reversible modification of lysine residues within the amino-terminal tail domains of core histones. Acetylation is mediated by histone acetyltransferase (HAT), which transfers acetyl groups from acetyl-coenzyme A to the lysine residue. In contrast, histone deacetylase (HDAC) acts in the reverse manner and removes the acetyl groups. Acetylation neutralizes the basic histone tails, weakens the histone-DNA interaction and, by promoting the accessibility of transcription factors to the gene locus, favors transcription. On the other hand, histone phosphorylation decreases transcription because, by adding negative charges, it seems to disrupt the interaction between histone tails and DNA. Several kinases critically involved in long-term memory formation, including ERK1/2, p38 MAP kinase, and ribosomal 6 kinase (RSK) are recruited in histone phosphoryation (228).
Once again, studies in Aplysia pioneered understanding of the molecular mechanisms underlying long-term plasticity and memory. Using the sensory motor co-culture system, it was found that long-term facilitation and long-term inhibition of the neurotransmitter release converge on the promoter of C/EBP, and it is the result of bidirectional modification of chromatin that leads, respectively, to gene activation or gene repression. Guan et al. (79) showed that facilitation is accompanied by induction of the expression of C/EBP after the activation of CREB1, which recruits CPB to acetylate histones at the C/EBP promoter. In contrast, inhibition of neurotransmitter release represses C/EBP by recruiting CREB2 and a Class II deacetylase (HDAC5) that deacetylate histones at the C/EBP promoter.
In mammals, using contextual fear conditioning, Levenson et al. (137) found that 1 h after fear conditioning there is a transient increase in the acetylation of histone H3, but not H4, in the CA1 region of the hippocampus. This increase depends on activation of the NMDA receptors and ERK. The functions of ERK, as well as that of mitogen- and stress-activated protein kinase (MSK1), a nuclear kinase downstream of ERK, are critical for histone acetylation and phosphorylation in the hippocampus after fear conditioning training, as is LTP (48). Furthermore LTP in hippocampal slices, as well as memory formation using fear conditioning consolidation, reconsolidation, and extinction learning, are enhanced by inhibitors of HDAC, indicating that, indeed, histone modifications have critical functions in long-term memory (37, 132, 137).
Do the histone modifications involved in memory formation affect gene expression globally or do they affect the expression of specific genes? Vecsey et al. (243) showed that the enhancement of hippocampus-dependent memory and hippocampal synaptic plasticity by HDAC inhibitors is mediated by CREB and the recruitment of CBP via the CREB-binding domain of the co-activator CBP. In agreement, the HDAC inhibitor trichostatin A does not globally alter gene expression; rather, it increases the expression of specific genes during memory consolidation. Moreover, MSK1 knock-out mice show impaired CREB phosphorylation and histone acetylation (48). Together, these results point to the conclusion that HDAC inhibitors enhance memory processes by selective activation of key genes regulated by the CREB-CBP complex.
Similar conclusions were reached on the basis of studies focusing on the function of CBP, a protein known to have intrinsic histone acetyltransferase activity. Heterologous mutations of CBP can cause Rubinstein-Taybi syndrome, a disorder characterized by growth and psychomotor delays as well as severe mental retardation (195). Dominant negative CBP transgenic mice that specifically inhibit HAT activity exhibit normal learning and short-term memory, but significant long-term memory deficits in the visual-paired comparison task and the spatial-task water maze, although they have normal contextual fear conditioning (126). Heterozygous cbp+/− mutant mice also show memory impairments in contextual fear conditioning but not in a water maze (7). Similarly, transgenic mice expressing a truncated form of CBP exhibit deficits in some but not other forms of LTP, as well as deficits in both water maze and contextual fear conditioning (256). Thus, despite differences in the forms of memory that are affected, which may reflect different requirements and brain regions involved in different tasks, these results indicate that CBP HAT activity has a critical function in long-term memory formation.
The pathway recruiting the transcription factor NF-kB has also been implicated in chromatin structure regulation during the post-retrieval phase of memory known as reconsolidation. Lubin and Sweatt (144) found that retrieval of contextual fear memories in rats activate the NF-kB pathway to regulate histone H3 phosphorylation and acetylation at specific gene promoters in the hippocampus and that this occurs through action of the kinase that phosphorylates the inhibitor kB (IKK). Inhibition of this activation blocks both memory reconsolidation and H3 modifications. It has also been reported that IKK can be co-recruited with CBP for gene regulation (18, 260).
In addition to chromatin changes, modifications of DNA have been found to be important for memory formation. For example, DNA (cytosine-5) methylation is critical for both synaptic plasticity and memory formation. Indeed the DNA within the promoters for the reelin and brain-derived neurotrophic factor genes, which have been implicated in the induction of synaptic plasticity in the adult hippocampus, undergo rapid and dramatic changes in cytosine methylation when DNA methyl transferase activity is blocked. Such blockage also disrupts the induction of LTP at Schaffer collateral synapses (139). In agreement with this finding, expression of DNA methyltransferases (DNMTs) is upregulated in the adult rat hippocampus after contextual fear conditioning and DNMT inhibition blocks memory formation. Fear conditioning is also associated with rapid methylation and transcriptional silencing of the memory suppressor gene protein phosphatase 1 (PP1), as well as demethylation and transcriptional activation of the synaptic plasticity gene reelin, indicating that both methyltransferase and demethylase activity occur during consolidation. Conversely, inhibition of DNMT activity after contextual fear conditioning learning blocks memory consolidation, probably as a consequence, at least in part, of modulation of histone acetylation (170). These results lead to the conclusion that DNA methylation is dynamically regulated in the adult nervous system and that this cellular mechanism is a crucial step in memory formation (169).
An intriguing and important finding is that DNA methylation of specific genes is also associated with early childhood experiences. These modifications are maintained during development. Specifically, patterns of methylation in the glucocorticoid receptor gene correlate with the quality of maternal care and are maintained into adulthood. Indeed the experience of having a good, nurturing mother correlates with an increase in hippocampal glucocorticoid receptor expression, altered histone acetylation, transcription factor NGFI-A binding to the glucocorticoid receptor promoter and, finally, a moderate response to stress in adulthood. This phenotype can be reversed by histone deacetylase inhibitors, suggesting a causal relation among epigenomic state, GR expression, and the maternal effect on stress responses in offspring. Thus, the epigenomic state of a gene can be established through behavioral programming and is potentially reversible (245).
IV. Conclusions
As a result of tremendous progress during the past 15 years, studies have led to the conclusion that the activation of patterns of gene expression organized in cascades, whereby transcription factors regulate the expression of regulatory immediate early genes, underlies long-term synaptic plasticity and long-term memory formation. Families of transcription factors involved include the CREB, C/EBP, Rel, AP-1 and NFkB families, suggesting that after learning information is transformed into long-term memory by inducing “different” cell states. Both transcription activators and repressors are recruited and long-term changes are accompanied by modifications of chromatin and DNA at genes known to be critically involved in long-term memory. Future investigations aim to identify the patterns of target genes, the expression regulation of which, over time, mediates long-term memory formation, as well as to elucidate the temporal dynamics of the gene regulation processes.
Acknowledgments
Many thanks to all the members of my laboratory for their invaluable contributions. Research in my laboratory was supported by NIMH grants R01 MH074736 and R01 MH65635, the Hirchl Foundation, and a NARSAD Independent Investigator Award.
References
- 1.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham WC, Dragunow M, Tate WP. The role of immediate early genes in the stabilization of long-term potentiation. Mol Neurobiol. 1991;5:297–314. doi: 10.1007/BF02935553. Review. [DOI] [PubMed] [Google Scholar]
- 3.Abraham WC, Mason SE, Demmer J, Williams JM, Richardson CL, Tate WP, Lawlor PA, Dragunow M. Correlations between immediate early gene induction and persistence of LTP. Neuroscience. 1993;56:717–727. doi: 10.1016/0306-4522(93)90369-q. [DOI] [PubMed] [Google Scholar]
- 4.Agranoff BW. Agents that block memory, in The Neurosciences: A study program. New York: Rockefeller University Press; 1967. [Google Scholar]
- 5.Ahmed T, Frey JU. Plasticity-specific phosphorylation of CaMKII, MAP-kinases and CREB during late-LTP in rat hippocampal slices in vitro. Neuropharmacology. 2005;49:477–492. doi: 10.1016/j.neuropharm.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 6.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 8.Albensi BC, Mattson MP. Evidence for the involvement of TNF and NF-kappaB in hippocampal synaptic plasticity. Synapse. 2000;35:151–159. doi: 10.1002/(SICI)1098-2396(200002)35:2<151::AID-SYN8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 9.Alberini CM, Ghirardi M, Metz R, Kandel ER. C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in Aplysia. Cell. 1994;76:1099–1114. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- 10.Alberini CM, Ghirardi M, Huang YY, Nguyen PV, Kandel ER. A molecular switch for the consolidation of long-term memory: cAMP-inducible gene expression. Ann N Y Acad Sci. 1995;758:261–286. doi: 10.1111/j.1749-6632.1995.tb24833.x. Review. [DOI] [PubMed] [Google Scholar]
- 11.Alberini CM. Genes to remember. J Exp Biol. 1999;202:2887–2891. doi: 10.1242/jeb.202.21.2887. [DOI] [PubMed] [Google Scholar]
- 12.Alberini CM. Mechanisms of memory stabilization: are consolidation and reconsolidation similar or distinct processes? Trends Neurosci. 2005;28:51–56. doi: 10.1016/j.tins.2004.11.001. Review. [DOI] [PubMed] [Google Scholar]
- 13.Alberini CM, Taubenfeld SM, Garcia-Osta A. CREB and the CREB-C/EBP-dependent Gene Expression Cascade in Long-term Memory. Cellscience reviews. 2005;Vol 2 http://www.cellscience.com/reviews6/CREB_long-term_memory.html. [Google Scholar]
- 14.Alberini CM. The role of protein synthesis during the labile phases of memory: revisiting the skepticism. Neurobiol Learn Mem. 2008;89:234–246. doi: 10.1016/j.nlm.2007.08.007. Epub 2007 Oct 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science; 2002. Chapter 7. [Google Scholar]
- 16.Alberts B, Bray D, Hopkin K, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Essential Cell Biology. 2/e. New York: Garland Science; 2004. Fig. 8–10. [Google Scholar]
- 17.Amin E, Pearce JM, Brown MW, Aggleton JP. Novel temporal configurations of stimuli produce discrete changes in immediate-early gene expression in the rat hippocampus. Eur J Neurosci. 2006;24:2611–2621. doi: 10.1111/j.1460-9568.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- 18.Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- 19.Anokhin KV, Rose SP. Learning-induced Increase of Immediate Early Gene Messenger RNA in the Chick Forebrain. Eur J Neurosci. 1991;3:162–167. doi: 10.1111/j.1460-9568.1991.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 20.Athos J, Impey S, Pineda VV, Chen X, Storm DR. Hippocampal CRE-mediated gene expression is required for contextual memory formation. Nat Neurosci. 2002;5:1119–1120. doi: 10.1038/nn951. [DOI] [PubMed] [Google Scholar]
- 21.Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci U S A. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balschun D, Wolfer DP, Gass P, Mantamadiotis T, Welzl H, Gunther S, Frey JU, Lipp HP. Does cAMP response element-binding protein have a pivotal role in hippocampal synaptic plasticity and hippocampus-dependent memory? J Neurosci. 2003;23:6304–6314. doi: 10.1523/JNEUROSCI.23-15-06304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barco A, Alarcon JM, Kandel ER. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell. 2002;108:689–703. doi: 10.1016/s0092-8674(02)00657-8. [DOI] [PubMed] [Google Scholar]
- 24.Barco A, Kandel ER. The Role of CREB and CBP in Brain Function. Weinheim: WILEY-VCH Verlag GmbH & Co.KGaA; 2006. p. 207. [Google Scholar]
- 25.Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- 26.Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95:211–223. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- 27.Barzilai A, Kennedy TE, Sweatt JD, Kandel ER. 5-HT modulates protein synthesis and the expression of specific proteins during long-term facilitation in Aplysia sensory neurons. Neuron. 1989;2:1577–1586. doi: 10.1016/0896-6273(89)90046-9. [DOI] [PubMed] [Google Scholar]
- 28.Bernabeu R, Bevilaqua L, Ardenghi P, Bromberg E, Schmitz P, Bianchin M, Izquierdo I, Medina JH. Involvement of hippocampal cAMP/cAMP-dependent protein kinase signaling pathways in a late memory consolidation phase of aversively motivated learning in rats. Proc Natl Acad Sci USA. 1997;94:7041–7046. doi: 10.1073/pnas.94.13.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertaina-Anglade V, Tramu G, Destrade C. Differential learning-stage dependent patterns of c-Fos protein expression in brain regions during the acquisition and memory consolidation of an operant task in mice. Eur J Neurosci. 2000;12:3803–3812. doi: 10.1046/j.1460-9568.2000.00258.x. [DOI] [PubMed] [Google Scholar]
- 30.Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- 31.Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Boccia M, Freudenthal R, Blake M, de la Fuente V, Acosta G, Baratti C, Romano A. Activation of hippocampal nuclear factor-kappa B by retrieval is required for memory reconsolidation. J Neurosci. 2007;27:13436–13445. doi: 10.1523/JNEUROSCI.4430-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolhuis JJ, Zijlstra GO, den Boer-Visser AM, Van der Zee EA. Localised neuronal activation in the zebra finch brain is related to the strength of song learning. Proc Natl Acad Sci USA. 2000;97:2282–2285. doi: 10.1073/pnas.030539097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourtchuladze R, Frenguelli B, Cioffi D, Blendy J, Schultz G, Silva AJ. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 35.Bozon B, Davis S, Laroche S. Regulated transcription of the immediate-early gene Zif268: mechanisms and gene dosage-dependent function in synaptic plasticity and memory formation. Hippocampus. 2002;12:570–577. doi: 10.1002/hipo.10100. Review. [DOI] [PubMed] [Google Scholar]
- 36.Brecht S, Simler S, Vergnes M, Mielke K, Marescaux C, Herdegen T. Repetitive electroconvulsive seizures induce activity of c-Jun N-terminal kinase and compartment-specific desensitization of c-Jun phosphorylation in the rat brain. Brain Res Mol Brain Res. 1999;68:101–108. doi: 10.1016/s0169-328x(99)00069-8. [DOI] [PubMed] [Google Scholar]
- 37.Bredy TW, Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn Mem. 2008;15:39–45. doi: 10.1101/lm.801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brightwell JJ, Smith CA, Countryman RA, Neve RL, Colombo PJ. Hippocampal overexpression of mutant creb blocks long-term, but not short-term memory for a socially transmitted food preference. Learn Mem. 2005;12:12–17. doi: 10.1101/lm.85005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brink JJ, Davis RE, Agranoff BW. Effects of puromycin, acetoxycycloheximide and actinomycin D on protein synthesis in goldfish brain. J Neurochem. 1966;13:889–896. doi: 10.1111/j.1471-4159.1966.tb10284.x. [DOI] [PubMed] [Google Scholar]
- 40.Burazin TCD, Grundlach AL. Rapid and transient increase in celllar immediate early gene and neuropeptide mRNAs in cortical and limbic areas after amygdaloid kindling seizures in the rat. Epilepsy Res. 1996;26:281–293. doi: 10.1016/s0920-1211(96)00060-5. [DOI] [PubMed] [Google Scholar]
- 41.Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 42.Carew TJ, Sutton MA. Molecular stepping stones in memory consolidation. Nat Neurosci. 2001;4:769–771. doi: 10.1038/90458. Review. [DOI] [PubMed] [Google Scholar]
- 43.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. Review. [DOI] [PubMed] [Google Scholar]
- 44.Castellucci VF, Frost WN, Goelet P, Montarolo PG, Schacher S, Morgan JA, Blumenfeld H, Kandel ER. Cell and molecular analysis of long-term sensitization in. Aplysia. J Physiol (Paris) 1986;81:349–357. [PubMed] [Google Scholar]
- 45.Chaisuksunt V, Campbell G, Zhang Y, Schachner M, Lieberman AR, Anderson PN. Expression of regeneration-related molecules in injured and regenerating striatal and nigral neurons. J Neurocytol. 2003;32:161–183. doi: 10.1023/b:neur.0000005601.59097.24. [DOI] [PubMed] [Google Scholar]
- 46.Chen A, Muzzio IA, Malleret G, Bartsch D, Verbitsky M, Pavlidis P, Yonan AL, Vronskaya S, Grody MB, Cepeda I, Gilliam TC, Kandel ER. Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron. 2003;39:655–669. doi: 10.1016/s0896-6273(03)00501-4. [DOI] [PubMed] [Google Scholar]
- 47.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 48.Chwang WB, Arthur JS, Schumacher A, Sweatt JD. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J Neurosci. 2007;27:12732–12742. doi: 10.1523/JNEUROSCI.2522-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cole AJ, Abu-Shakra S, Saffen DW, Baraban JM, Worley PF. Rapid rise in transcription factor mRNAs in rat brain after electroshock-induced seizures. J Neurochem. 1990;55:1920–1927. doi: 10.1111/j.1471-4159.1990.tb05777.x. [DOI] [PubMed] [Google Scholar]
- 50.Countryman RA, Orlowski JD, Brightwell JJ, Oskowitz AZ, Colombo PJ. CREB phosphorylation and c-Fos expression in the hippocampus of rats during acquisition and recall of a socially transmitted food preference. Hippocampus. 2005;15:56–67. doi: 10.1002/hipo.20030. [DOI] [PubMed] [Google Scholar]
- 51.Da Silva WC, Bonini JS, Bevilaqua LR, Medina JH, Izquierdo I, Cammarota M. Inhibition of mRNA synthesis in the hippocampus impairs consolidation and reconsolidation of spatial memory. Hippocampus. 2008;18:29–39. doi: 10.1002/hipo.20362. [DOI] [PubMed] [Google Scholar]
- 52.Dash PK, Hochner B, Kandel ER. Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature. 1990;345:718–721. doi: 10.1038/345718a0. [DOI] [PubMed] [Google Scholar]
- 53.Davis HP, Squire LR. Protein synthesis and memory: a review. Psychol Bull. 1984;96:518–559. [PubMed] [Google Scholar]
- 54.Davis S, Bozon B, Laroche S. How necessary is the activation of the immediate early gene zif268 in synaptic plasticity and learning? Behav Brain Res. 2003;142:17–30. doi: 10.1016/s0166-4328(02)00421-7. [DOI] [PubMed] [Google Scholar]
- 55.De Cesare D, Sassone-Corsi P. Transcriptional regulation by cyclic AMP-responsive factors. Prog Nucleic Acid Res Mol Biol. 2000;64:343–369. doi: 10.1016/s0079-6603(00)64009-6. Review. [DOI] [PubMed] [Google Scholar]
- 56.Donahue CP, Jensen RV, Ochiishi T, Eisenstein I, Zhao M, Shors T, Kosik KS. Transcriptional profiling reveals regulated genes in the hippocampus during memory formation. Hippocampus. 2002;12:821–833. doi: 10.1002/hipo.10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dragunow M, Currie RW, Faull RL, Robertson HA, Jansen K. Immediate-early genes, kindling and long-term potentiation. Neurosci Biobehav Rev. 1989;13:301–313. doi: 10.1016/s0149-7634(89)80066-1. Review. [DOI] [PubMed] [Google Scholar]
- 58.Dragunow M, Faull RL, Waldvogel HJ, Williams MN, Leah J. Elevated expression of jun and fos-related proteins in transplanted striatal neurons. Brain Res. 1991;558:321–324. doi: 10.1016/0006-8993(91)90785-t. [DOI] [PubMed] [Google Scholar]
- 59.Dragunow M. A role for immediate early transcription factors in learning and memory. Behav Genetics. 1996;20:293–299. doi: 10.1007/BF02359385. [DOI] [PubMed] [Google Scholar]
- 60.Dragunow M. CREB and neurodegeneration. Front Biosci. 2004;9:100–103. doi: 10.2741/1197. [DOI] [PubMed] [Google Scholar]
- 61.Dudai Y, Eisenberg M. Rites of passage of the engram: reconsolidation and the lingering consolidation hypothesis. Neuron. 2004;44:93–100. doi: 10.1016/j.neuron.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 62.Falls WA, Kogan JH, Silva AJ, Willott JF, Carlson S, Turner JG. Fear-potentiated startle, but not prepulse inhibition of startle, is impaired in CREBalphadelta−/− mutant mice. Behav Neurosci. 2000;114:998–1004. doi: 10.1037//0735-7044.114.5.998. [DOI] [PubMed] [Google Scholar]
- 63.Freeman FM, Rose SP. Expression of Fos and Jun proteins following passive avoidance training in the day-old chick. Learn Mem. 1999;6:389–397. [PMC free article] [PubMed] [Google Scholar]
- 64.Freeman WM, Nader MA, Nader SH, Robertson DJ, Gioia L, Mitchell SM, Daunais JB, Porrino LJ, Friedman DP, Vrana KE. Chronic cocaine-mediated changes in non-human primate nucleus accumbens gene expression. J Neurochem. 2001;77:542–549. doi: 10.1046/j.1471-4159.2001.00252.x. [DOI] [PubMed] [Google Scholar]
- 65.French PJ, O'Connor V, Jones MW, Davis S, Errington ML, Voss K, Truchet B, Wotjak C, Stean T, Doyère V, Maroun M, Laroche S, Bliss TV. Subfield-specific immediate early gene expression associated with hippocampal long-term potentiation in vivo. Eur J Neurosci. 2001;13:968–976. doi: 10.1046/j.0953-816x.2001.01467.x. [DOI] [PubMed] [Google Scholar]
- 66.Freudenthal R, Locatelli F, Hermitte G, Maldonado H, Lafourcade C, Delorenzi A, Romano A. Kappa-B like DNA-binding activity is enhanced after spaced training that induces long-term memory in the crab Chasmagnathus. Neurosci Lett. 1998;242:143–146. doi: 10.1016/s0304-3940(98)00059-7. [DOI] [PubMed] [Google Scholar]
- 67.Freudenthal R, Romano A. Participation of Rel/NF-kappaB transcription factors in long-term memory in the crab Chasmagnathus. Brain Res. 2000;855:274–281. doi: 10.1016/s0006-8993(99)02358-6. [DOI] [PubMed] [Google Scholar]
- 68.Freudenthal R, Romano A, Routtenberg A. Transcription factor NF-kappaB activation after in vivo perforant path LTP in mouse hippocampus. Hippocampus. 2004;14:677–683. doi: 10.1002/hipo.20020. [DOI] [PubMed] [Google Scholar]
- 69.Freudenthal R, Boccia MM, Acosta GB, Blake MG, Merlo E, Baratti CM, Romano A. NF-kappaB transcription factor is required for inhibitory avoidance long-term memory in mice. Eur J Neurosci. 2005;21:2845–2852. doi: 10.1111/j.1460-9568.2005.04126.x. [DOI] [PubMed] [Google Scholar]
- 70.Frey U, Krug M, Reymann KG, Matthies H. Anisomycin, an inhibitor of protein synthesis, blocks late phases of LTP phenomena in the hippocampal CA1 region in vitro. Brain Res. 1988;452:57–65. doi: 10.1016/0006-8993(88)90008-x. [DOI] [PubMed] [Google Scholar]
- 71.Frey U, Frey S, Schollmeier F, Krug M. Influence of actinomycin D a RNA synthesis inhibitor, on long-term potentiation in rat hippocampal neurons in vivo and in vitro. J Physiol. 1996;490:703–711. doi: 10.1113/jphysiol.1996.sp021179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frey JU. Long-lasting hippocampal plasticity: cellular model from memory consolidation? Cell polarity and subcellular RNA localization. In: Richter D, editor. Results and problems in cell differentiation. Vol. 34. Berlin, Heidelberg: Springer-Verlag; 2001. pp. 27–40. [DOI] [PubMed] [Google Scholar]
- 73.Friedman AD. C/EBPalpha induces PU.1 and interacts with AP-1 and NF-kappaB to regulate myeloid development. Blood Cells Mol Dis. 2007;39:340–343. doi: 10.1016/j.bcmd.2007.06.010. Epub 2007 Jul 31. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gall C, Lauterborn J, Isackson P, White J. Seizures, neuropeptide regulation, and mRNA expression in the hippocampus. Prog Brain Res. 1990;83:371–390. doi: 10.1016/s0079-6123(08)61263-7. Review. [DOI] [PubMed] [Google Scholar]
- 75.Gass P, Herdegen T, Bravo R, Kiessling M. Spatiotemporal induction of immediate early genes in the rat brain after limbic seizures: effects of NMDA receptor antagonist MK-801. Eur J Neurosci. 1993;5:933–943. doi: 10.1111/j.1460-9568.1993.tb00944.x. [DOI] [PubMed] [Google Scholar]
- 76.Gelderblom M, Eminel S, Herdegen T, Waetzig V. c-Jun N-terminal kinases (JNKs) and the cytoskeleton--functions beyond neurodegeneration. Int J Dev Neurosci. 2004;22:559–564. doi: 10.1016/j.ijdevneu.2004.07.014. Review. [DOI] [PubMed] [Google Scholar]
- 77.Goelet P, Castellucci VF, Schacher S, Kandel ER. The long and the short of long-term memory--a molecular framework. Nature. 1986;322:419–422. doi: 10.1038/322419a0. [DOI] [PubMed] [Google Scholar]
- 78.Gold PE. Protein synthesis inhibition and memory: formation vs amnesia. Neurobiol Learn Mem. 2008;89:201–211. doi: 10.1016/j.nlm.2007.10.006. Epub 2007 Dec 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, Kandel ER. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- 80.Guzowski JF, McGaugh JL. Anti-sense oligodeoxynucleotide-mediated disruption of hippocampal CREB protein levels impairs memory of a spatial task. Proc Natl Acad Sci USA. 1997;94:2693–2698. doi: 10.1073/pnas.94.6.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J Neurosci. 2001;21:5089–5098. doi: 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gräff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behav Brain Res. 2008 Feb 12; doi: 10.1016/j.bbr.2008.01.021. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 83.Hai T and Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 84.Hall J, Thomas KL, Everitt BJ. Rapid and selective induction of BDNF expression in the hippocampus during contextual learning. Nat Neurosci. 2000;3:533–535. doi: 10.1038/75698. [DOI] [PubMed] [Google Scholar]
- 85.Hall J, Thomas KL, Everitt BJ. Cellular imaging of zif268 expression in the hippocampus and amygdala during contextual and cued fear memory retrieval: selective activation of hippocampal CA1 neurons during recall of contextual memories. J Neurosci. 2001;21:2186–2193. doi: 10.1523/JNEUROSCI.21-06-02186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiology and Molecular Biology Reviews. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, Neve RL, Guzowski JF, Silva AJ, Josselyn SA. Neuronal competition and selection during memory formation. Science. 2007;316:457–460. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- 88.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 89.Hayward MD, Duman RS, Nestler EJ. Induction of the c-fos proto-oncogene during opiate withdrawal in the locus coeruleus and other regions of rat brain. Brain Res. 1990;525:256–266. doi: 10.1016/0006-8993(90)90872-9. [DOI] [PubMed] [Google Scholar]
- 90.He J, Yamada K, Nakajima A, Kamei H, Nabeshima T. Learning and memory in two different reward tasks in a radial arm maze in rats. Behav Brain Res. 2002;134:139–148. doi: 10.1016/s0166-4328(01)00460-0. [DOI] [PubMed] [Google Scholar]
- 91.Helmstetter FJ, Parsons RG, Gafford GM. Macromolecular synthesis, distributed synaptic plasticity, and fear conditioning. Neurobiol Learn Mem. 2008;89:324–337. doi: 10.1016/j.nlm.2007.09.002. Epub 2007 Oct 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Herdegen T, Tölle TR, Bravo R, Zieglgänsberger W, Zimmermann M. Sequential expression of JUN B, JUN D and FOS B proteins in rat spinal neurons: cascade of transcriptional operations during nociception. Neurosci Lett. 1991a;129:221–224. doi: 10.1016/0304-3940(91)90466-7. [DOI] [PubMed] [Google Scholar]
- 93.Herdegen T, Leah JD, Manisali A, Bravo R, Zimmermann M. c-JUN-like immunoreactivity in the CNS of the adult rat: basal and transynaptically induced expression of an immediate-early gene. Neuroscience. 1991b;41:643–654. doi: 10.1016/0306-4522(91)90356-s. [DOI] [PubMed] [Google Scholar]
- 94.Herdegen T, Kummer W, Fiallos CE, Leah J, Bravo R. Expression of c-JUN, JUN B and JUN D proteins in rat nervous system following transection of vagus nerve and cervical sympathetic trunk. Neuroscience. 1991c;45:413–422. doi: 10.1016/0306-4522(91)90237-i. [DOI] [PubMed] [Google Scholar]
- 95.Herdegen T, Leah JD. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res Brain Res Rev. 1998;28:370–490. doi: 10.1016/s0165-0173(98)00018-6. Review. [DOI] [PubMed] [Google Scholar]
- 96.Hernandez PJ, Abel T. The role of protein synthesis in memory consolidation: progress amid decades of debate. Neurobiol Learn Mem. 2008;89:293–311. doi: 10.1016/j.nlm.2007.09.010. Epub 2007 Nov 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Heurteaux C, Messier C, Destrade C, Lazdunski M. Memory processing and apamin induce immediate early gene expression in mouse brain. Brain Res Mol Brain Res. 1993a;18:17–22. doi: 10.1016/0169-328x(93)90169-p. [DOI] [PubMed] [Google Scholar]
- 98.Heurteaux C, Bertaina V, Widmann C, Lazdunski M. K+ channel openers prevent global ischemia-induced expression of c-fos, c-jun, heat shock protein, and amyloid beta-protein precursor genes and neuronal death in rat hippocampus. Proc Natl Acad Sci U S A. 1993b;90:9431–9435. doi: 10.1073/pnas.90.20.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heynen AJ, Bear MF. Long-term potentiation of thalamocortical transmission in the adult visual cortex in vivo. J Neurosci. 2001;21:9801–9813. doi: 10.1523/JNEUROSCI.21-24-09801.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Honjo K, Furukubo-Tokunagua K. Induction of cAMP response element-binding protein-dependent medium-term memory by appetitive gustatory reinforcement in Drosophila larvae. J Neurosci. 2005;25:7905–7913. doi: 10.1523/JNEUROSCI.2135-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Igaz LM, Vianna MR, Medina JH, Izquierdo I. Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear-motivated learning. J Neurosci. 2002;22:6781–6789. doi: 10.1523/JNEUROSCI.22-15-06781.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Impey S, Mark M, Villacres EC, Poser S, Chavkin C, Storm DR. Induction of CRE-mediated gene expression by stimuli that generate long-lasting LTP in area CA1 of the hippocampus. Neuron. 1996;16:973–982. doi: 10.1016/s0896-6273(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 103.Jamieson AC, Miller JC, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat Rev Drug Discov. 2003;2:361–368. doi: 10.1038/nrd1087. Review. [DOI] [PubMed] [Google Scholar]
- 104.Jarvis ED, Mello CV, Nottebohm F. Associative learning and stimulus novelty influence the song-induced expression of an immediate early gene in the canary forebrain. Learn Mem. 1995;2:62–80. doi: 10.1101/lm.2.2.62. [DOI] [PubMed] [Google Scholar]
- 105.Jarvis ED, Ribeiro S, da Silva ML, Ventura D, Veilliard J, Mello CV. Behaviourally driven gene expression reveals song nuclei in hummingbird brain. Nature. 2000;406:628–632. doi: 10.1038/35020570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jenkins R, Hunt SP. Long-term increase in the levels of c-jun mRNA and jun protein-like immunoreactivity in motor and sensory neurons following axon damage. Neurosci Lett. 1991;129:107–110. doi: 10.1016/0304-3940(91)90731-8. [DOI] [PubMed] [Google Scholar]
- 107.Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004;16:1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 108.Jones MW, French PJ. TVP Bliss and K Rosenblum, Molecular mechanisms of long-term potentiation in the insular cortex in vivo. J Neurosci. 1999;19:RC36. doi: 10.1523/JNEUROSCI.19-21-j0002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and the consolidation of long-term memories. Nat Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- 110.Josselyn SA, Kida S, Silva AJ. Inducible repression of CREB function disrupts amygdala-dependent memory. Neurobiol Learn Mem. 2004;82:159–163. doi: 10.1016/j.nlm.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 111.Josselyn SA, Nguyen PV. CREB, synapses and memory disorders: past progress and future challenges. Curr Drug Targets CNS Neurol Disord. 2005;4:481–497. doi: 10.2174/156800705774322058. Review. [DOI] [PubMed] [Google Scholar]
- 112.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. Review. [DOI] [PubMed] [Google Scholar]
- 113.Kaltschmidt B, Mikenberg I, Widera D, Kaltschmidt C. In: Transcription Factors in the Nervous System: Development, Brain Function, and Diseases. Thiel Gerald., editor. Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA; 2006. p. 329. [Google Scholar]
- 114.Kaltschmidt B, Ndiaye D, Korte M, Pothion S, Arbibe L, Prüllage M, Pfeiffer J, Lindecke A, Staiger V, Israël A, Kaltschmidt C, Mémet S. NF-kappaB regulates spatial memory formation and synaptic plasticity through protein kinase A/CREB signaling. Mol Cell Biol. 2006;26:2936–2946. doi: 10.1128/MCB.26.8.2936-2946.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kaminska B, Filipkowski RK, Zurkowska G, Lason W, Przewlocki R, Kaczmarek L. Dynamic changes in the composition of the AP-1 transcription factor DNA-binding activity in rat brain following kainate-induced seizures and cell death. Eur J Neurosci. 1994;6:1558–1566. doi: 10.1111/j.1460-9568.1994.tb00546.x. [DOI] [PubMed] [Google Scholar]
- 116.Kandel ER. Genes, nerve cells, and the remembrance of things past. J Neuropsychiatry Clin Neurosci. 1989;1:103–125. doi: 10.1176/jnp.1.2.103. Review. [DOI] [PubMed] [Google Scholar]
- 117.Kandel ER, Jessel T, Schwartz J. Principles of neural science. IV Edition. McGraw Hill; 2000. [Google Scholar]
- 118.Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 119.Kassed CA, Willing AE, Garbuzova-Davis S, Sanberg PR, Pennypacker KR. Lack of NF-kappaB p50 exacerbates degeneration of hippocampal neurons after chemical exposure and impairs learning. Exp Neurol. 2002;176:277–288. doi: 10.1006/exnr.2002.7967. [DOI] [PubMed] [Google Scholar]
- 120.Kassed CA, Herkenham M. NF-kappaB p50-deficient mice show reduced anxiety-like behaviors in tests of exploratory drive and anxiety. Behav Brain Res. 2004;154:577–584. doi: 10.1016/j.bbr.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 121.Kindy MS, Carney JP, Dempsey RJ, Carney JM. Ischemic induction of protooncogene expression in gerbil brain. J Mol Neurosci. 1991;2:217–228. [PubMed] [Google Scholar]
- 122.Klann E, Sweatt JD. Altered protein synthesis is a trigger for long-term memory formation. Neurobiol Learn Mem. 2008;89:247–259. doi: 10.1016/j.nlm.2007.08.009. Epub 2007 Oct 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Knapska E, Kaczmarek L. A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog Neurobiol. 2004;74:183–211. doi: 10.1016/j.pneurobio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 124.Kockel L, Zeitlinger J, Staszewski LM, Mlodzik M, Bohmann D. Jun in Drosophila development: redundant and nonredundant functions and regulation by two MAPK signal transduction pathways. Genes Dev. 1997;11:1748–1758. doi: 10.1101/gad.11.13.1748. Erratum in: Genes Dev 12: 447, 1998. [DOI] [PubMed] [Google Scholar]
- 125.Kogan JH, Frankland PW, Silva AJ. Long-term memory underlying hippocampus-dependent social recognition in mice. Hippocampus. 2000;10:47–56. doi: 10.1002/(SICI)1098-1063(2000)10:1<47::AID-HIPO5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 126.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kovács KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, Cardinaux JR. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc Natl Acad Sci U S A. 2007;104:4700–4705. doi: 10.1073/pnas.0607524104. Epub 2007 Mar 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lamprecht R, Dudai Y. Transient expression of c-Fos in rat amygdala during training is required for encoding conditioned taste aversion memory. Learn Mem. 1996;3:31–41. doi: 10.1101/lm.3.1.31. [DOI] [PubMed] [Google Scholar]
- 129.Lamprecht R, Hazvi S, Dudai Y. cAMP response element-binding protein in the amygdala is required for long-but not short-term conditioned taste aversion memory. J Neurosci. 1997;17:8443–8450. doi: 10.1523/JNEUROSCI.17-21-08443.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lanaud P, Maggio R, Gale K, Grayson DR. Temporal and spatial patterns of expression of c-fos, zif/268, c-jun and jun-B mRNAs in rat brain following seizures evoked focally from the deep prepiriform cortex. Exp Neurol. 1993;119:20–31. doi: 10.1006/exnr.1993.1003. [DOI] [PubMed] [Google Scholar]
- 131.Lane MD, Tang QQ, Jiang MS. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem Biophys Res Commun. 1999;266:677–683. doi: 10.1006/bbrc.1999.1885. Review. [DOI] [PubMed] [Google Scholar]
- 132.Lattal KM, Barrett RM, Wood MA. Systemic or intrahippocampal delivery of histone deacetylase inhibitors facilitates fear extinction. Behav Neurosci. 2007;121:1125–1131. doi: 10.1037/0735-7044.121.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. Review. [DOI] [PubMed] [Google Scholar]
- 134.LeDoux J. The Emotional Brain: The Mysterious Underpinnings of Emotional Life. Simon & Schuster; New York: 1996. [Google Scholar]
- 135.Lee JA, Kim HK, Kim KH, Han JH, Lee YS, Lim CS, Chang DJ, Kubo T, Kaang BK. Overexpression of and RNA interference with the CCAAT enhancer-binding protein on long-term facilitation of Aplysia sensory to motor synapses. Learn Mem. 2001;8:220–226. doi: 10.1101/lm.40201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Leutgeb JK, Frey JU, Behnisch T. Single cell analysis of activity-dependent cyclic AMP-responsive element-binding protein phosphorylation during long-lasting long-term potentiation in area CA1 of mature rat hippocampal-organotypic cultures. Neuroscience. 2005;131:601–610. doi: 10.1016/j.neuroscience.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 137.Levenson JM, Choi S, Lee SY, Cao YA, Ahn HJ, Worley KC, Pizzi M, Liou HC, Sweatt JD. A bioinformatics analysis of memory consolidation reveals involvement of the transcription factor c-rel. J Neurosci. 2004;24:3933–3943. doi: 10.1523/JNEUROSCI.5646-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. Review. [DOI] [PubMed] [Google Scholar]
- 139.Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 140.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 141.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. Review. Erratum in: Nat Rev Immunol 2: 975, 2002. [DOI] [PubMed] [Google Scholar]
- 142.Liu RY, Fioravante D, Shah S, Byrne JH. cAMP response element-binding protein 1 feedback loop is necessary for consolidation of long-term synaptic facilitation in Aplysia. J Neurosci. 2008;28:1970–1976. doi: 10.1523/JNEUROSCI.3848-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. Review. [DOI] [PubMed] [Google Scholar]
- 144.Lubin FD, Sweatt JD. The IkappaB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron. 2007;55:942–957. doi: 10.1016/j.neuron.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lynch MA. Long-term potentiation and memory. Physiol Rev. 2004;84:87–136. doi: 10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- 146.Mack KJ, Cortner J, Mack P and Farnham PJ. Krox-20 messenger RNA and protein expression in the adult central nervous system. Mol Brain Res. 1992;14:117–123. doi: 10.1016/0169-328x(92)90018-7. [DOI] [PubMed] [Google Scholar]
- 147.Malkani S, Rosen JB. Specific induction of early growth response gene 1 in the lateral nucleus of the amygdala following contextual fear conditioning in rats. Neuroscience. 2000;97:693–702. doi: 10.1016/s0306-4522(00)00058-0. [DOI] [PubMed] [Google Scholar]
- 148.Martin KC, Michael D, Rose JC, Barad M, Casadio A, Zhu H, Kandel ER. MAP kinase translocates into the nucleus of the presynaptic cell and is required for long-term facilitation in Aplysia . Neuron. 1997;18:899–912. doi: 10.1016/s0896-6273(00)80330-x. [DOI] [PubMed] [Google Scholar]
- 149.Mataga N, Fujishima S, Condi BG, Hensch TK. Experience-dependent plasticity of mouse visual cortex in the absence of the neuronal activity-dependent marker egr1 zif268. J Neurosci. 2001;21:9724–9732. doi: 10.1523/JNEUROSCI.21-24-09724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Matsuo R, Murayama A, Saitoh Y, Sakaki Y, Inokuchi K. Identification and cataloging of genes induced by long-lasting long-term potentiation in awake rats. J Neurochem. 2000;74:2239–2249. doi: 10.1046/j.1471-4159.2000.0742239.x. [DOI] [PubMed] [Google Scholar]
- 151.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 152.McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. doi: 10.1038/sj.npp.1301544. [DOI] [PubMed] [Google Scholar]
- 153.McGaugh JL. Memory--a century of consolidation. Science. 2000;287:248–251. doi: 10.1126/science.287.5451.248. Review. [DOI] [PubMed] [Google Scholar]
- 154.McKnight SL. McBindall--a better name for CCAAT/enhancer binding proteins? Cell. 2001;107:259–261. doi: 10.1016/s0092-8674(01)00543-8. Review. [DOI] [PubMed] [Google Scholar]
- 155.Meberg PJ, Kinney WR, Valcourt EG, Routtenberg A. Gene expression of the transcription factor NF-kappa B in hippocampus: regulation by synaptic activity. Brain Res Mol Brain Res. 1996;38:179–190. doi: 10.1016/0169-328x(95)00229-l. [DOI] [PubMed] [Google Scholar]
- 156.Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. Epub 2003 Aug 31. Erratum in: Nat Neurosci 6:1329, 2003. [DOI] [PubMed] [Google Scholar]
- 157.Meffert MK, Baltimore D. Physiological functions for brain NF-kappaB. Trends Neurosci. 2005;28:37–43. doi: 10.1016/j.tins.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 158.Mello CV, Vicario DS, Clayton DF. Song presentation induces gene expression in the songbird forebrain. Proc Natl Acad Sci USA. 1992;89:6818–6822. doi: 10.1073/pnas.89.15.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mello CV, Clayton DF. Song-induced ZENK gene expression in auditory pathways of songbird brain and its relation to the song control system. J Neurosci. 1994;14:6652–6666. doi: 10.1523/JNEUROSCI.14-11-06652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mello CV, Velho TA, Pinaud R. Song-induced gene expression: a window on song auditory processing and perception. Ann N Y Acad Sci. 2004;1016:263–281. doi: 10.1196/annals.1298.021. [DOI] [PubMed] [Google Scholar]
- 161.Mellström B, Achaval M, Montero D, Naranjo JR, Sassone-Corsi P. Differential expression of the jun family members in rat brain. Oncogene. 1991;6:1959–1964. [PubMed] [Google Scholar]
- 162.Merlo E, Freudenthal R, Romano A. The IkappaB kinase inhibitor sulfasalazine impairs long-term memory in the crab Chasmagnathus. Neuroscience. 2002;112:161–172. doi: 10.1016/s0306-4522(02)00049-0. [DOI] [PubMed] [Google Scholar]
- 163.Merlo E, Freudenthal R, Maldonado H, Romano A. Activation of the transcription factor NF-kappaB by retrieval is required for long-term memory reconsolidation. Learn Mem. 2005;12:23–29. doi: 10.1101/lm.82705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Metz R, Ziff E. cAMP stimulates the C/EBP-related transcription factor rNFIL-6 to trans-locate to the nucleus and induce c-fos transcription. Genes Dev. 1991;5:1754–1766. doi: 10.1101/gad.5.10.1754. [DOI] [PubMed] [Google Scholar]
- 165.Meyer TE, Habener JF. Cyclic adenosine 3',5'-monophosphate response element binding protein (CREB) and related transcription-activating deoxyribonucleic acid-binding proteins. Endocr Rev. 1993;14:269–290. doi: 10.1210/edrv-14-3-269. [DOI] [PubMed] [Google Scholar]
- 166.Michael D, Martin KC, Seger R, Ning MM, Baston R, Kandel ER. Repeated pulses of serotonin required for long-term facilitation activate mitogen-activated protein kinase in sensory neurons of Aplysia . Proc Natl Acad Sci U S A. 1998;95:1864–1869. doi: 10.1073/pnas.95.4.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mikenberg I, Widera D, Kaus A, Kaltschmidt B, Kaltschmidt C. Transcription factor NF-kappaB is transported to the nucleus via cytoplasmic dynein/dynactin motor complex in hippocampal neurons. PLoS ONE. 2007 Jul 11;2(7):e589. doi: 10.1371/journal.pone.0000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mileusnic R, Anokhin K, Rose SP. Antisense oligodeoxynucleotides to c-fos are amnestic for passive avoidance in the chick. Neuroreport. 1996;7:1269–1272. doi: 10.1097/00001756-199605170-00010. [DOI] [PubMed] [Google Scholar]
- 169.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 170.Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. Epub 2007 Sep 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mioduszewska B, Jaworski J, Kaczmarek L. Inducible cAMP early repressor (ICER) in the nervous system--a transcriptional regulator of neuronal plasticity and programmed cell death. J Neurochem. 2003;87:1313–1320. doi: 10.1046/j.1471-4159.2003.02116.x. Review. [DOI] [PubMed] [Google Scholar]
- 172.Mohamed HA, Yao W, Fioravante D, Smolen PD, Byrne JH. cAMP-response elements in Aplysia creb1, creb2, and Ap-uch promoters: implications for feedback loops modulating long term memory. J Biol Chem. 2005;280:27035–27043. doi: 10.1074/jbc.M502541200. Epub 2005 May 11. [DOI] [PubMed] [Google Scholar]
- 173.Montarolo PG, Goelet P, Castellucci VF, Morgan J, Kandel ER, Schacher S. Science. 1986;234:1249–1254. doi: 10.1126/science.3775383. [DOI] [PubMed] [Google Scholar]
- 174.Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 175.Montminy MR, Gonzalez GA, Yamamoto KK. Characteristics of the cAMP response unit. Metabolism. 1990;39:6–12. doi: 10.1016/0026-0495(90)90198-l. [DOI] [PubMed] [Google Scholar]
- 176.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. Review. [DOI] [PubMed] [Google Scholar]
- 177.Montminy M, Koo SH, Zhang X. The CREB family: key regulators of hepatic metabolism. Ann Endocrinol (Paris) 2004;65:73–75. doi: 10.1016/s0003-4266(04)95634-x. [DOI] [PubMed] [Google Scholar]
- 178.Morgan JI, Curran T. Stimulus-transcription coupling in neurons: role of cellular immediate-early genes. Trends Neurosci. 1989;12:459–462. doi: 10.1016/0166-2236(89)90096-9. Review. [DOI] [PubMed] [Google Scholar]
- 179.Morrow BA, Elsworth JD, Inglis FM, Roth RH. An antisense oligonucleotide reverses the footshock-induced expression of fos in the rat medial prefrontal cortex and the subsequent expression of conditioned fear-induced immobility. J Neurosci. 1999;19:5666–5673. doi: 10.1523/JNEUROSCI.19-13-05666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nader K, Schafe GE, LeDoux JE. The labile nature of consolidation theory. Nat Rev Neurosci. 2000;1:216–219. doi: 10.1038/35044580. Review. [DOI] [PubMed] [Google Scholar]
- 181.Naranjo JR, Mellström B, Achaval M, Sassone-Corsi P. Molecular pathways of pain: Fos/Jun-mediated activation of a noncanonical AP-1 site in the prodynorphin gene. Neuron. 1991;6:607–617. doi: 10.1016/0896-6273(91)90063-6. [DOI] [PubMed] [Google Scholar]
- 182.Neale JH, Klinger PD, Agranoff BW. Camptothecin blocks memory of conditioned avoidance in the goldfish. Science. 1973;179:1243–1246. doi: 10.1126/science.179.4079.1243. [DOI] [PubMed] [Google Scholar]
- 183.Nestler EJ. Cellular responses to chronic treatment with drugs of abuse. Crit Rev Neurobiol. 1993;7:23–39. Review. [PubMed] [Google Scholar]
- 184.Nicoll RA, Malenka RC. Expression mechanisms underlying NMDA receptor-dependent long-term potentiation. Ann N Y Acad Sci. 1999;868:515–525. doi: 10.1111/j.1749-6632.1999.tb11320.x. [DOI] [PubMed] [Google Scholar]
- 185.Okuno H, Miyashita Y. Expression of the transcription factor Zif268 in the temporal cortex of monkeys during visual paired associate learning. Eur J Neurosci. 1996;8:2118–2128. doi: 10.1111/j.1460-9568.1996.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 186.O'Neill LA, Kaltschmidt C. NF-kappa B: a crucial transcription factor for glial and neuronal cell function. Trends Neurosci. 1997;20:252–258. doi: 10.1016/s0166-2236(96)01035-1. [DOI] [PubMed] [Google Scholar]
- 187.Orphanides G, LaGrange T, Reinberg D. The general initiation factors of RNA polymerase II. Genes and Development. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 188.Osten P, Valsamis L, Harris A, Sacktor TC. Protein synthesis-dependent formation of protein kinase Mzeta in long-term potentiation. J Neurosci. 1996;16:2444–2451. doi: 10.1523/JNEUROSCI.16-08-02444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Panomics AP-1 Activation Pathway. 2007 AP-1 Reporter 293 Stable Cell Line: How It Works http://www.panomics.com/AP1stablecellline.cfm,
- 190.Papa M, Pellicano MP, Welzl H, Sadile AG. Distributed changes in c-Fos and c-Jun immunoreactivity in the rat brain associated with arousal and habituation to novelty. Brain Res Bull. 1993;32:509–515. doi: 10.1016/0361-9230(93)90299-q. [DOI] [PubMed] [Google Scholar]
- 191.Parsons RG, Gafford GM, Baruch DE, Riedner BA, Helmstetter FJ. Long-term stability of fear memory depends on the synthesis of protein but not mRNA in the amygdala. Eur J Neurosci. 2006;23:1853–1859. doi: 10.1111/j.1460-9568.2006.04723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pedreira ME, Dimant B, Maldonado H. Inhibitors of protein and RNA synthesis block context memory and long-term habituation in the crab Chasmagnathus. Pharmacol Biochem Behav. 1996;54:611–617. doi: 10.1016/0091-3057(95)02206-6. [DOI] [PubMed] [Google Scholar]
- 193.Perazzona B, Isabel G, Preat T, Davis RL. The role of cAMP response element-binding protein in Drosophila long-term memory. J Neurosci. 2004;24:8823–8828. doi: 10.1523/JNEUROSCI.4542-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Persengiev SP, Green MR. The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis. 2003;8:225–228. doi: 10.1023/a:1023633704132. Review. [DOI] [PubMed] [Google Scholar]
- 195.Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ, Breuning MH. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 196.Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S, Kandel ER. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron. 2002;34:447–462. doi: 10.1016/s0896-6273(02)00684-0. [DOI] [PubMed] [Google Scholar]
- 197.Quevedo J, Vianna MR, Roesler R, de-Paris F, Izquierdo I, Rose SP. Two time windows of anisomycin-induced amnesia for inhibitory avoidance training in rats: protection from amnesia by pretraining but not pre-exposure to the task apparatus. Learn Mem. 1999;6:600–607. doi: 10.1101/lm.6.6.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Rainbow TC. Role of RNA and protein synthesis in memory formation. Neurochem Res. 1979;4:297–312. doi: 10.1007/BF00963800. [DOI] [PubMed] [Google Scholar]
- 199.Raivich G, Bohatschek M, Da Costa C, Iwata O, Galiano M, Hristova M, Nateri AS, Makwana M, Riera-Sans L, Wolfer DP, Lipp HP, Aguzzi A, Wagner EF, Behrens A. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 200.Raivich G, Behrens A. Role of the AP-1 transcription factor c-Jun in developing, adult and injured brain. Prog Neurobiol. 2006;78:347–363. doi: 10.1016/j.pneurobio.2006.03.006. Epub 2006 May 22. [DOI] [PubMed] [Google Scholar]
- 201.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002;365:561–575. doi: 10.1042/BJ20020508. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Richter-Levin G, Thomas KL, Hunt SP, Bliss TVP. Dissociation between genes activated in long-term potentiation and in spatial learning in the rat. Neurosci Lett. 1988;251:41–44. doi: 10.1016/s0304-3940(98)00476-5. [DOI] [PubMed] [Google Scholar]
- 203.Richardson CL, Tate WP, Mason SE, Lawlor PA, Dragunow M, Abraham WC. Correlation between the induction of an immediate early gene, zif/268, and long-term potentiation in the dentate gyrus. Brain Res. 1992;580:147–154. doi: 10.1016/0006-8993(92)90938-6. [DOI] [PubMed] [Google Scholar]
- 204.Roeder RG, Rutter WJ. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969;224:234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- 205.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends in Biochemical Science. 1996;21:327–335. [PubMed] [Google Scholar]
- 206.Roesler WJ. The role of C/EBP in nutrient and hormonal regulation of gene expression. Annu Rev Nutr. 2001;21:141–165. doi: 10.1146/annurev.nutr.21.1.141. Review. [DOI] [PubMed] [Google Scholar]
- 207.Romano A, Freudenthal R, Merlo E, Routtenberg A. Evolutionarily-conserved role of the NF-kappaB transcription factor in neural plasticity and memory. Eur J Neurosci. 2006;24:1507–1516. doi: 10.1111/j.1460-9568.2006.05022.x. [DOI] [PubMed] [Google Scholar]
- 208.Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 209.Rosen JB, Fanselow MS, Young SL, Sitcoske M, Maren S. Immediate-early gene expression in the amygdala following footshock stress and contextual fear conditioning. Brain Res. 1998;796:132–142. doi: 10.1016/s0006-8993(98)00294-7. [DOI] [PubMed] [Google Scholar]
- 210.Rössler OG, Stefano L, Bauer I, Thiel Gerald. Stimulus-Transcription Coupling in the Nervous System: The Zinc Finger Protein Egr-1. Weinheim: WILEY-VCH Verlag GmbH & Co.KGaA; 2006. p. 379. [Google Scholar]
- 211.Sangha S, Scheibenstock A, Lukowiak K. Reconsolidation of a long-term memory in Lymnaea requires new protein and RNA synthesis and the soma of right pedal dorsal 1. J Neurosci. 2003;23:8034–8040. doi: 10.1523/JNEUROSCI.23-22-08034.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sanyal S, Sandstrom DJ, Hoeffer CA, Ramaswami M. AP-1 functions upstream of CREB to control synaptic plasticity in Drosophila . Nature. 2002;416:870–874. doi: 10.1038/416870a. [DOI] [PubMed] [Google Scholar]
- 213.Sara SJ. Retrieval and reconsolidation: toward a neurobiology of remembering. Learn Mem. 2000;7:73–84. doi: 10.1101/lm.7.2.73. Review. [DOI] [PubMed] [Google Scholar]
- 214.Schrem H, Klempnauer J, Borlak J. Liver-enriched transcription factors in liver function and development. Part II: the C/EBPs and D site-binding protein in cell cycle control, carcinogenesis, circadian gene regulation, liver regeneration, apoptosis, and liver-specific gene regulation. Pharmacol Rev. 2004;56:291–330. doi: 10.1124/pr.56.2.5. Review. [DOI] [PubMed] [Google Scholar]
- 215.Schulz DJ, Goaillard JM, Marder EE. Quantitative expression profiling of identified neurons reveals cell-specific constraints on highly variable levels of gene expression. Proc Natl Acad Sci U S A. 2007;104:13187–13191. doi: 10.1073/pnas.0705827104. Epub 2007 Jul 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Schulz S, Siemer H, Krug M, Hollt V. Direct evidence for biphasic cAMP responsive element-binding protein phosphorylation during long-term potentiation in the rat dentate gyrus in vivo. J Neurosci. 1999;19:5683–5692. doi: 10.1523/JNEUROSCI.19-13-05683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Segal M, Murphy DD. CREB activation mediates plasticity in cultured hippocampal neurons. Neural Plast. 1998;6:1–7. doi: 10.1155/NP.1998.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sigurdsson T, Doyère V, Cain CK, LeDoux JE. Long-term potentiation in the amygdala: a cellular mechanism of fear learning and memory. Neuropharmacology. 2007;52:215–227. doi: 10.1016/j.neuropharm.2006.06.022. Epub 2006 Aug 21. [DOI] [PubMed] [Google Scholar]
- 219.Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and Memory. Ann Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- 220.Smith CA, Countryman RA, Sahuque LL, Colombo PJ. Time-courses of Fos expression in rat hippocampus and neocortex following acquisition and recall of a socially transmitted food preference. Neurobiol Learn Mem. 2007;88:65–74. doi: 10.1016/j.nlm.2007.03.001. Epub 2007 Apr 19. [DOI] [PubMed] [Google Scholar]
- 221.Sonnenberg JL, Macgregor-Leon PF, Curran T, Morgan JI. Dynamic alterations occur in the levels and composition of transcription factor AP-1 complexes after seizure. Neuron. 1989;3:359–365. doi: 10.1016/0896-6273(89)90260-2. [DOI] [PubMed] [Google Scholar]
- 222.Squire L, Barondes S. Actinomycin-D:effects on memory at different times after training. Nature. 1970;225:649–650. doi: 10.1038/225649a0. [DOI] [PubMed] [Google Scholar]
- 223.Squire LR. Memory and brain. New York: Oxford; 1987. [Google Scholar]
- 224.Stanciu M, Radulovic J, Spiess J. Phosphorylated cAMP response element binding protein in the mouse brain after fear conditioning: relationship to Fos production. Brain Res Mol Brain Res. 2001;94:15–24. doi: 10.1016/s0169-328x(01)00174-7. [DOI] [PubMed] [Google Scholar]
- 225.Sterneck E, Paylor R, Jackson-Lewis V, Libbey M, Przedborski S, Tessarollo L, Crawley JN, Johnson PF. Selectively enhanced contextual fear conditioning in mice lacking the transcriptional regulator CCAAT/enhancer binding protein delta. Proc Natl Acad Sci U S A. 1998;95:10908–10913. doi: 10.1073/pnas.95.18.10908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. Epub 2005 Dec 20. Erratum in: Nat Neurosci 9: 292, 2006. [DOI] [PubMed] [Google Scholar]
- 227.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 228.Taniura H, Sng JC, Yoneda Y. Histone modifications in the brain. Neurochem Int. 2007;51:85–91. doi: 10.1016/j.neuint.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 229.Taubenfeld SM, Wiig KA, Bear MF, Alberini CM. A molecular correlate of memory and amnesia in the hippocampus. Nat Neurosci. 1999;2:309–310. doi: 10.1038/7217. [DOI] [PubMed] [Google Scholar]
- 230.Taubenfeld SM, Wiig KA, Monti B, Dolan B, Pollonini G, Alberini CM. Fornix-dependent induction of hippocampal CCAAT enhancer-binding protein β and δ co-localizes with phosphorylated cAMP response element binding protein and accompanies long-term memory consolidation. J Neurosci. 2001a;21:84–91. doi: 10.1523/JNEUROSCI.21-01-00084.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Taubenfeld SM, Milekic MH, Monti B, Alberini CM. The consolidation of new but not reactivated memory requires hippocampal C/EBPbeta. Nat Neurosci. 2001b;4:813–818. doi: 10.1038/90520. [DOI] [PubMed] [Google Scholar]
- 232.Taubenfeld SM, Stevens KA, Pollonini G, Ruggiero J, Alberini CM. Profound molecular changes following hippocampal slice preparation: loss of AMPA receptor subunits and uncoupled mRNA/protein expression. J Neurochem. 2002;81:1348–1360. doi: 10.1046/j.1471-4159.2002.00936.x. [DOI] [PubMed] [Google Scholar]
- 233.Thiel G, editor. Transcription factors in the nervous system. Wiley-VCH; 2006. [Google Scholar]
- 234.Thomas KL, Hall J, Everitt BJ. Cellular imaging with zif268 expression in the rat nucleus accumbens and frontal cortex further dissociates the neural pathways activated following the retrieval of contextual and cued fear memory. Eur J Neurosci. 2002;16:1789–1796. doi: 10.1046/j.1460-9568.2002.02247.x. [DOI] [PubMed] [Google Scholar]
- 235.Thut PD, Lindell TJ. Alpha-amanitin inhibition of mouse brain form II ribonucleic acid polymerase and passive avoidance retention. Mol Pharmacol. 1974;10:146–154. [PubMed] [Google Scholar]
- 236.Tinsley MR, Quinn JJ, Fanselow MS. The role of muscarinic and nicotinic cholinergic neurotransmission in aversive conditioning: comparing pavlovian fear conditioning and inhibitory avoidance. Learn Mem. 2004;11:35–42. doi: 10.1101/lm.70204. Review. [DOI] [PubMed] [Google Scholar]
- 237.Tischmeyer W, Grimm R, Schicknick H, Brysch W, Schlingensiepen KH. Sequence-specific impairment of learning by c-jun antisense oligonucleotides. Neuroreport. 1994;5:1501–1504. doi: 10.1097/00001756-199407000-00023. [DOI] [PubMed] [Google Scholar]
- 238.Tischmeyer W, Grimm R. Activation of immediate early genes and memory formation. Cell Mol Life Sci. 1999;55:564–574. doi: 10.1007/s000180050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Tokuyama W, Okuno H, Hashimoto T, Li YX, Miyashita Y. Selective zif268 mRNA induction in the perirhinal cortex of macaque monkeys during formation of visual pair-association memory. J Neurochem. 2002;81:60–70. doi: 10.1046/j.1471-4159.2002.00790.x. [DOI] [PubMed] [Google Scholar]
- 240.Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- 241.Tsokas P, Grace EA, Chan P, Ma T, Sealfon SC, Iyengar R, Landau EM, Blitzer RD. Local protein synthesis mediates a rapid increase in dendritic elongation factor 1A after induction of late long-term potentiation. J Neurosci. 2005;25:5833–5843. doi: 10.1523/JNEUROSCI.0599-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ubeda M, Wang XZ, Zinszner H, Wu I, Habener JF, Ron D. Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol Cell Biol. 1996;16:1479–1489. doi: 10.1128/mcb.16.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Warburton EC, Glover CP, Massey PV, Wan H, Johnson B, Bienemann A, Deuschle U, Kew JN, Aggleton JP, Bashir ZI, Uney J, Brown MW. cAMP responsive element-binding protein phosphorylation is necessary for perirhinal long-term potentiation and recognition memory. J Neurosci. 2005;27:6296–6303. doi: 10.1523/JNEUROSCI.0506-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. Epub 2004 Jun 27. [DOI] [PubMed] [Google Scholar]
- 246.Wedel A, Ziegler-Heitbrock HW. The C/EBP family of transcription factors. Immunobiology. 1995;193:171–185. doi: 10.1016/s0171-2985(11)80541-3. Review. [DOI] [PubMed] [Google Scholar]
- 247.Weitemier AZ, Ryabinin AE. Subregion-specific differences in hippocampal activity between Delay and Trace fear conditioning: an immunohistochemical analysis. Brain Res. 2004;995:55–65. doi: 10.1016/j.brainres.2003.09.054. [DOI] [PubMed] [Google Scholar]
- 248.Wellmann H, Kaltschmidt B, Kaltschmidt C. Retrograde transport of transcription factor NF-kappa B in living neurons. J Biol Chem. 2001;276:11821–11829. doi: 10.1074/jbc.M009253200. [DOI] [PubMed] [Google Scholar]
- 249.Wessel TC, Joh TH, Volpe BT. In situ hybridization analysis of c-fos and c-jun expression in the rat brain following transient forebrain ischemia. Brain Res. 1991;567:231–240. doi: 10.1016/0006-8993(91)90800-b. [DOI] [PubMed] [Google Scholar]
- 250.Wetzel W, Ott T, Matthies H. Is actinomycin D suitable for the investigation of memory processes? Pharmacol Biochem Behav. 1976;4:515–519. doi: 10.1016/0091-3057(76)90190-8. [DOI] [PubMed] [Google Scholar]
- 251.White JD, Gall CM. Differential regulation of neuropeptide and proto-oncogene mRNA content in the hippocampus following recurrent seizures. Brain Res. 1987;427:21–29. doi: 10.1016/0169-328x(87)90040-4. [DOI] [PubMed] [Google Scholar]
- 252.Williams J, Dragunow M, Lawlor P, Mason S, Abraham WC, Leah J, Bravo R, Demmer J, Tate W. Krox20 may play a key role in the stabilisation of long-term potentiation. Mol Brain Res. 1995;28:87–93. doi: 10.1016/0169-328x(94)00187-j. [DOI] [PubMed] [Google Scholar]
- 253.Williams SC, Cantwell CA, Johnson PF. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 1991;5:1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 254.Winston SM, Hayward MD, Nestler EJ, Duman RS. Chronic electroconvulsive seizures down-regulate expression of the immediate-early genes c-fos and c-jun in rat cerebral cortex. J Neurochem. 1990;54:1920–1925. doi: 10.1111/j.1471-4159.1990.tb04892.x. [DOI] [PubMed] [Google Scholar]
- 255.Wisden W, Errington ML, Williams S, Dunnett SB, Waters C, Hitchcock D, Evan G, Bliss TV, Hunt SP. Differential expression of immediate early genes in the hippocampus and spinal cord. Neuron. 1990;4:603–614. doi: 10.1016/0896-6273(90)90118-y. [DOI] [PubMed] [Google Scholar]
- 256.Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Worley PF, Bhat RV, Baraban JM, Erickson CA, McNaughton BL, Barnes CA. Thresholds for synaptic activation of transcription factors in hippocampus: correlation with long-term enhancement. J Neurosci. 1993;13:4776–4786. doi: 10.1523/JNEUROSCI.13-11-04776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Yamagata K, Kaufmann WE, Lanahan A, Papapavlou M, Barnes CA, Andreasson KI, Worley PF. Egr-3/Pilot Egr-1, a zinc finger transcription factor, is rapidly regulated by activity in brain neurons and colocalizes with Egr-1/zif268. Learn Mem. 1994;1:140–152. [PubMed] [Google Scholar]
- 259.Yamamoto N, Hegde AN, Chain DG, Schwartz JH. Activation and degradation of the transcription factor C/EBP during long-term facilitation in. Aplysia. J Neurochem. 1999;73:2415–2423. doi: 10.1046/j.1471-4159.1999.0732415.x. [DOI] [PubMed] [Google Scholar]
- 260.Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- 261.Yeh SH, Lin CH, Lee CF, Gean PW. A requirement of nuclear factor-kappaB activation in fear-potentiated startle. J Biol Chem. 2002;277:46720–46729. doi: 10.1074/jbc.M206258200. [DOI] [PubMed] [Google Scholar]
- 262.Yin JCP, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant-negative CREB transgene specifically blocks long-term memory in Drosophila melanogaster. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 263.Yoneda Y, Ogita K, Kabutoz H, Mori A. Selectively high expression of the transcription factor AP1 in telencephalic structures of epileptic E1 mice. Neurosci Lett. 1993;161:161–164. doi: 10.1016/0304-3940(93)90284-r. [DOI] [PubMed] [Google Scholar]
- 264.Yount GL, Ponsalle P, White JD. Pentylenetetrazole-induced seizures stimulate transcription of early and late response genes. Brain Res Mol Brain Res. 1994;21:219–224. doi: 10.1016/0169-328x(94)90252-6. [DOI] [PubMed] [Google Scholar]
- 265.Yuan Q, Harley CW, Darby-King A, Neve RL, McLean JH. Early odor preference learning in the rat: bidirectional effects of cAMP response element-binding protein (CREB) and mutant CREB support a causal role for phosphorylated CREB. J Neurosci. 2003;23:4760–4765. doi: 10.1523/JNEUROSCI.23-11-04760.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Yukawa K, Tanaka T, Tsuji S, Akira S. Expressions of CCAAT/Enhancer-binding proteins beta and delta and their activities are intensified by cAMP signaling as well as Ca2+/calmodulin kinases activation in hippocampal neurons. J Biol Chem. 1998;273:31345–31351. doi: 10.1074/jbc.273.47.31345. [DOI] [PubMed] [Google Scholar]
- 267.Zawel L, Reinberg D. Initiation of transcription by RNA polymerase II: A multi-step process. Progress in Nucleic Acids Research. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]
- 268.Zhang JJ, Okutani F, Inoue S, Kaba H. Activation of the cyclic AMP response element-binding protein signaling pathway in the olfactory bulb is required for the acquisition of olfactory aversive learning in young rats. Neuroscience. 2003;117:707–713. doi: 10.1016/s0306-4522(02)00962-4. [DOI] [PubMed] [Google Scholar]