Developmental Fate and Cellular Maturity Encoded in Human Regulatory DNA Landscapes (original) (raw)
. Author manuscript; available in PMC: 2014 Mar 21.
SUMMARY
Cellular-state information between generations of developing cells may be propagated via regulatory regions. We report consistent patterns of gain and loss of DNase I-hypersensitive sites (DHSs) as cells progress from embryonic stem cells (ESCs) to terminal fates. DHS patterns alone convey rich information about cell fate and lineage relationships distinct from information conveyed by gene expression. Developing cells share a proportion of their DHS landscapes with ESCs; that proportion decreases continuously in each cell type as differentiation progresses, providing a quantitative benchmark of developmental maturity. Developmentally stable DHSs densely encode binding sites for transcription factors involved in autoregulatory feedback circuits. In contrast to normal cells, cancer cells extensively reactivate silenced ESC DHSs and those from developmental programs external to the cell lineage from which the malignancy derives. Our results point to changes in regulatory DNA landscapes as quantitative indicators of cell-fate transitions, lineage relationships, and dysfunction.
INTRODUCTION
Under natural conditions, tissue and cellular differentiation along defined lineages is characterized by an inexorably forward-moving process that terminates in highly specialized cells. Waddington, following Morgan (Morgan, 1901), characterized the process of development as essentially “epigenetic” (from “epigenesis”) (Waddington, 1939) and also introduced the metaphor of an “epigenetic landscape” (Waddington, 1940), which he depicted with a ball rolling down a hill of bifurcating valleys symbolizing the specification of defined cell lineages and fates during the progress of differentiation (Waddington, 1939, 1957). It is notable that Waddington's usage of epigenetic to denote the origination and propagation of information about cellular states during differentiation differs considerably from its recent reformulation to mean “on the genome” and its association with chemical modifications to DNA or chromatin (Ptashne, 2007). Here we employ the classical usage throughout.
Waddington astutely reasoned that epigenesis is a “historical” process requiring a memory “faculty” to keep directed lineage programs on track (Waddington, 1939). Indeed, developing cells are frequently exposed to stimuli, whether exogenous (e.g., a morphogen) or endogenous (e.g., a transcription factor [TF]), that can permanently alter cellular fate. Whether or in what form cells in fact maintain information concerning prior developmental fate decisions during epigenesis is currently unknown.
The epigenetic landscape paradigm has also been invoked to explain abnormal processes such as oncogenesis (Pujadas and Feinberg, 2012). Cancer cells are widely described as being “de-differentiated” compared with their normal counterparts, based on limited analyses of metabolic (Warburg, 1956), histological (Gleason and Mellinger, 1974), gene-activity (Hirszfeld et al., 1932; Tatarinov, 1964), and proliferative and self-renewal phenotypes (Beard, 1902; Waddington, 1935). However, quantifying this concept and generalizing it beyond a few selected markers have proven difficult.
Chromatin structure represents a highly plastic vehicle for specifying cellular regulatory states and is a conceptually attractive template for recording and transmitting epigenetic information (Bernstein et al., 2006; Hawkins et al., 2010; Paige et al., 2012; Wamstad et al., 2012; Zhu et al., 2013). DNase I-hypersensitive sites (DHSs) represent focal alterations in the primary structure of chromatin that result from engagement of sequence-specific transcription factors in place of a canonical nucleosome (Gross and Garrard, 1988; Thurman et al., 2012). In a classic experiment, Groudine and Weintraub demonstrated that induced DHSs could be propagated to, and stably perpetuated by, daughter cells even after the inducing stimulus had been withdrawn (Groudine and Weintraub, 1982). This result suggests that newly arising DHSs created by TF occupancy of quiescent regulatory DNA have the potential to encode cellular states and to perpetuate that information through continued TF occupancy in daughter cells. Whether, or to what extent, such a mechanism operates during normal development and differentiation, however, is currently unknown.
To explore the role of TF-driven chromatin structure at regulatory DNA in normal and transformed cells during epigenesis, we analyzed genome-wide patterns of DHSs across a wide array of cell types and states, including definitive adult primary cells, embryonic stem cells (ESCs), cells undergoing directed lineage differentiation from ESCs to cardiomyocytes, and diverse cancer cell types. Our findings, detailed below, are interpreted to indicate four fundamental conclusions. First, patterns of DHSs in definitive cells encode “memory” of early developmental fate decisions that establish lineage hierarchies. Second, lineage differentiation couples the extensive activation of novel regulatory DNA compartments with propagation and sequential restriction of the ES DHS landscape as a function of cellular maturity. Third, developmentally stable DHSs chiefly encode binding sites for self-regulating TFs, suggesting a mechanistic role for TF-encoded feedback circuits in propagating developmental information. Finally, oncogenesis is accompanied by a disordered retrograde remodeling of the regulatory DNA landscape in a fashion that defies normal developmental pathways and departs fundamentally from the paradigm of the epigenetic landscape. Together these findings indicate a central role for patterning and propagation of regulatory DNA marked by DHSs in the genesis and proper maintenance of developmental programs.
RESULTS
Lineage Programming of Human Regulatory DNA
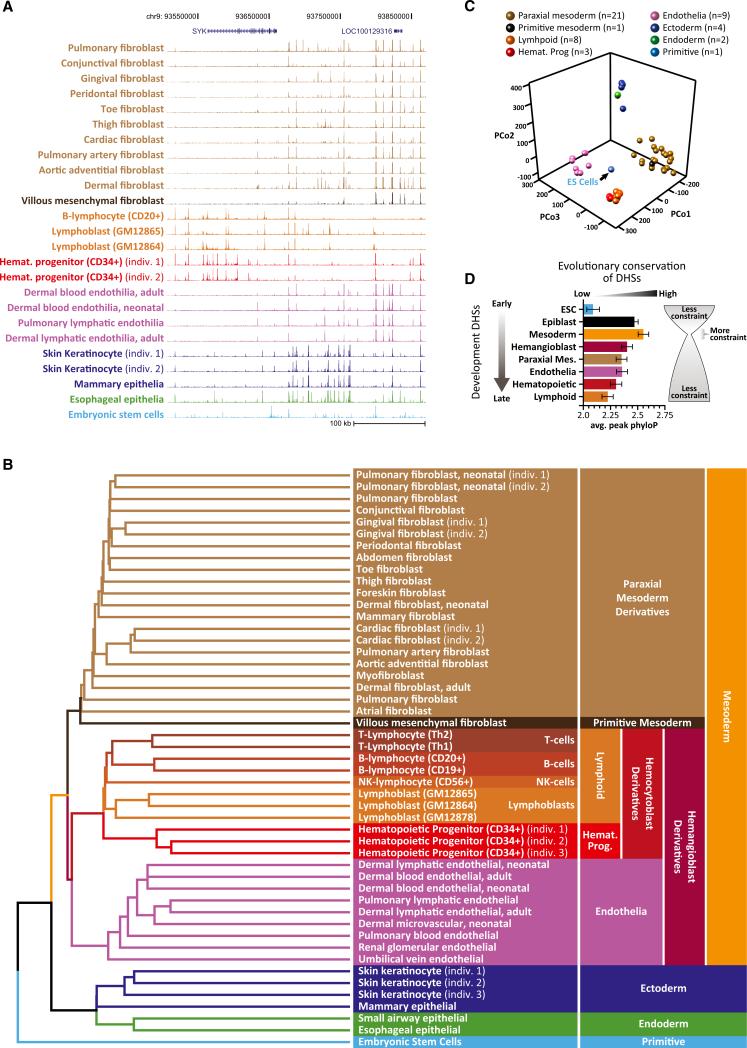
Regulatory DNA landscapes defined by DHSs are both highly cell type specific and highly stable (Thurman et al., 2012). We first sought to determine how the regulatory landscapes of diverse definitive cells were related to one another and to the regulatory DNA of ESCs. To address this, we collected genome-wide maps of DHSs from human ESCs plus 38 diverse normal definitive primary cell types (Thurman et al., 2012) for which anatomical and histological origins could be unambiguously verified. To expand the phenotypic range of cell types and to deepen coverage of the well-characterized hematopoietic lineage, we obtained nine additional definitive cell samples from adult donors, including B cells (CD19+, CD20+), natural killer (NK) cells (CD56+), CD34+ hematopoietic progenitors (three separate donors), and skin keratinocytes (three donors). The relative representation of different major embryological lineages (mesoderm, ectoderm, endoderm) among these 49 cell types parallels that of recognized cell types (Bard et al., 2005), of which those of mesodermal origin comprise the significant majority. We performed DNase I-hypersensitivity mapping on each of the 49 cell types using a common protocol and delineated DHSs using a common algorithm that has been extensively validated for both sensitivity and specificity (John et al., 2011; Thurman et al., 2012) (Experimental Procedures), resulting in an average of 161,160 autosomal DHSs per cell type (at false discovery rate [FDR] 1%, range 91,720 to 257,172; Table S1). Although most DHSs were highly cell selective, preliminary inspection of the DNase I profiles suggested systematic commonalities between major cell-type groups (Figures 1A, S1A, and S1B).
Figure 1. Lineage Programming of Human Regulatory DNA.
(A) Evidence of lineage patterning in primary DHS data. DNase I cleavage-density profiles for 24 exemplary primary human cell types and ESCs across an ~350 kb region along chromosome 9. Cell types are colored according to their embryological derivation as indicated in (B).
(B) Clustering DHS profiles recovers precise embryological relationships. Unbiased clustering of the linear patterning of DHSs from 48 diverse, definitive cell types plus ESCs. Branches and cell types are colored according to their embryological origin, with embryological ancestors common to multiple cell types indicated on the right. Note the rooting of the tree by ESCs and the partitioning of major branches corresponding to the trilaminar embryo. Note also the demarcation of early fate decisions such as partitioning of hemangioblast derivatives into endothelia and blood.
(C) PCoA of cell-type relationships. Shown is each cell type from (B) projected into a three-dimensional principal coordinates space. Cell-type coloring is indicated above. Note the centrality of ESCs and the spatial separation of major lineage groups.
(D) “Hourglass” pattern of regulatory DNA conservation across the developmental spectrum. Shown is the mean evolutionary conservation (phyloP, x axis) of DHSs common to the indicated lineage branches. Error bars represent 95% confidence intervals.
See also Figure S1 and S3 and Tables S1 and S2.
To visualize these relationships quantitatively, we considered each DHS to be either present or absent within a given cell type (versus total DNase I signal, to avoid biasing toward promoters, which display higher average DNase I sensitivity than distal elements) (Thurman et al., 2012) and computed the Euclidean distance between all nonredundant pairs of cell types. Rendering the results with simple unsupervised nearest-neighbor clustering (Figure 1B) produced an ab initio dendrogram that recapitulated known cell-lineage relationships with remarkable detail, as well as broader features of embryological origin. On a gross level, ESCs occupied the deepest root, and derivatives of the three germ layers (mesoderm, ectoderm, and endoderm) were correctly partitioned into separate high-level clusters (Figure 1B). Mesodermal progeny were further partitioned into paraxial mesoderm, primitive mesoderm, and hemangioblast derivatives. The common embryological origin of endothelia and blood was clearly represented, as was the fine partitioning of the hematopoietic tree into hematopoietic progenitors, lymphoid and myeloid cells, and the different subtypes of lymphoid tissue, including B cells, T cells, NK cells, and more primitive lymphoblastoid cells. Although relationships between the derivatives of paraxial mesoderm are less well understood, we observed subgroups that were organized into anatomical units, such as grouping of heart and great vessel stroma. The distinctiveness of these major cluster groups was clearly evinced by displaying the aforementioned pairwise Euclidean distance measures with a three-dimensional principal coordinate analysis (PCoA) (Figure 1C). This analysis also revealed that the regulatory DNA landscape of ESCs occupies a central position relative to all other cell types.
To confirm the robustness of the clustering, we performed bootstrap analysis, which determined the nearly complete stability of all major branches (Figure S1C). The robustness of the clustered cell relationships was further attested bythe strict cohesion of multiple samples of the same cell type, including gingival fibroblasts (n = 2), cardiac fibroblasts (n = 2), hematopoietic progenitors (n = 3), and keratinocytes (n = 3), that were derived from different individuals at different times. In addition, we prospectively tested the ability of the dendrogram to classify eight additional cell types of diverse embryological origin (Figures S1D–S1F). Importantly, clustering gene-expression patterns for the same cell types failed to recover the fundamental lineage-branching relationships exposed by clustering DHS patterns (Figures S3A and S3B), including rooting of the lineage tree in ESCs, and showed improper high-level segregation of germ-layer derivatives and improper partitioning of mesodermal derivatives. These results demonstrate that the dendrogram in Figure 1B is not driven by functional convergence on gene-expression patterns.
The fact that the aforementioned lineage relationships—including representation of specific primitive commitment events—can be derived from a simple clustering of the DHS landscapes of terminally differentiated cells suggests that the linear patterning of regulatory DNA along the genome encodes an imprint of prior cellular fate decisions. Given that ESCs represent a common developmental ancestor to the other cell types, the centrality of ESCs within the PCoA plot suggests that significant yet distinct components of the ESC regulatory landscape are shared in each of the definitive cell types (see below).
An “Hourglass” Pattern of Conservation at Developmental Regulatory DNA
Next, we sought to determine whether the pattern of developmental maturity reflected in the dendrogram was systematically paralleled by patterns of evolutionary constraint on regulatory DNA. We first identified regulatory DNA stably arising at seven distinct inferred developmental branch points (epiblast, mesoderm, hemangioblast, paraxial mesoderm, endothelia, hematopoietic, and lymphoid) by identifying DHSs common to the corresponding dependent branches of the dendrogram in Figure 1. We then used phyloP to calculate the mean level of evolutionary constraint for each set of elements (Experimental Procedures). This analysis revealed that regulatory DNA common to mesodermal derivatives (and thus inferred to be stably arising during the onset of the mesodermal lineage) is significantly more evolutionarily constrained than that arising during either early embryogenesis or later lineage differentiation (Figure 1D). This pattern is compatible with the “hourglass” model of development (Duboule, 1994; Raff, 1996) that has been variably described using cross-species morphology (Von Baer, 1828), gene expression (Kalinka et al., 2010), and gene conservation (Domazet-Lošo and Tautz, 2010).
Developmental Persistence of Chromatin Accessibility at Primitive Enhancers
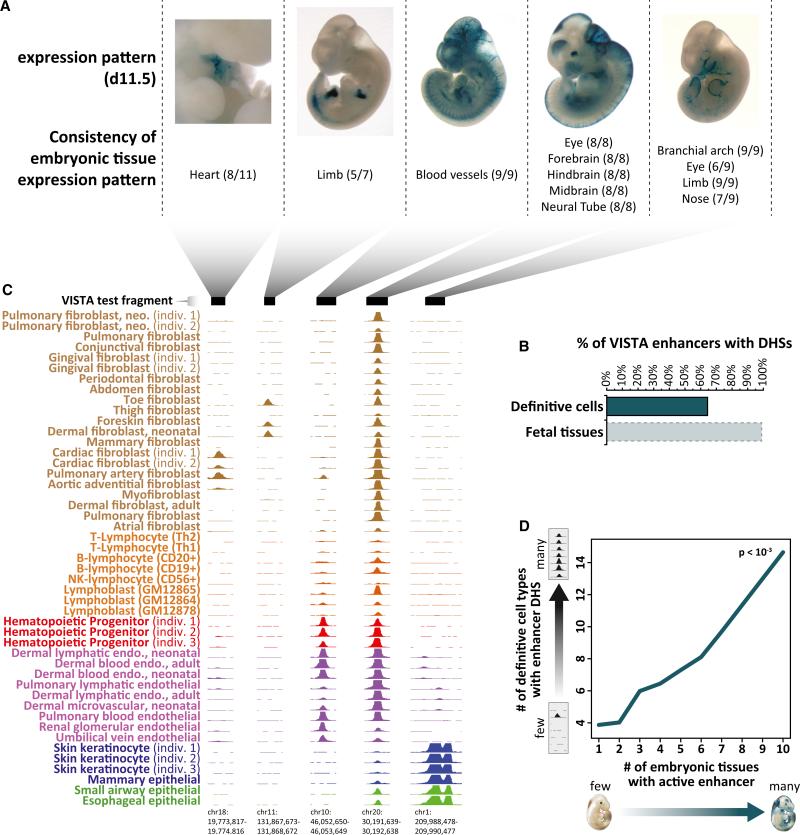
We next asked whether enhancers active during early development could be persistently marked by DHSs in definitive cells. Systematic studies of evolutionarily conserved human DNA elements in transgenic mice have identified >700 early developmental enhancers (Pennacchio et al., 2006), each of which displays consistent activity in one or more embryonic tissues (Figure 2A). Of 721 nonpromoter human enhancers with reproducible tissue-staining patterns in transgenic day 11.5 embryos, a surprising proportion—64%—exhibit DNase I hypersensitivity in at least one definitive human cell type (Figure 2B). To quantify the tissue activity spectra of these elements, we systematically collated images of enhancer-driven lacZ expression in individual transgenic animals and related these with cross-cell-type patterns of DNase I hypersensitivity at the same elements in definitive cells. For example, an enhancer that is selectively active in embryonic heart tissue (Figure 2A, 1st image) is DNase I hypersensitive selectively within cells derived from human heart and great vessel structures (Figure 2C), and an enhancer that is selectively active in embryonic blood vessels (Figure 2A, 3rd image) is DNase I hypersensitive selectively within hemangioblast derivatives (endothelia and hematopoietic progenitors; Figure 2C). By contrast, an enhancer with extremely broad tissue activity (Figure 2A, 4th image) is DNase I hypersensitive in nearly all definitive cell types (Figure 2C).
Figure 2. Developmental Persistence of Chromatin Accessibility at Primitive Enhancers.
(A) Mouse day 11.5 embryonic tissue activity (blue lacZ staining) of five representative transgenic human enhancer elements from the VISTA database. Shown below each image are numbers of individual embryos with enhancer activity (staining) in the indicated anatomical structure.
(B) Persistence of DNase I hypersensitivity at embryonic enhancers. The percentage of embryonic enhancers from the VISTA database marked with DHSs in one or more definitive cell types (top) or early human fetal tissues (~day 70–150) (bottom).
(C) DNase I hypersensitivity at five enhancer elements corresponding to (A) across 47 definitive cell types from Figure 1. Note the relationship between the anatomical staining patterns in (A) and the cellular restriction (or lack thereof) of DNase I hypersensitivity.
(D) Embryonic enhancer tissue spectrum parallels DHS spectrum in definitive cells. The number of embryonic tissues with an active enhancer by lacZ staining (x axis) is linearly proportional (linear regression p value < 10–3) to the number of definitive cell types showing DNase I hypersensitivity at the same enhancer (y axis).
See also Figure S2.
These findings generalize across the spectrum of enhancers: 100% of enhancers active in embryonic blood vessels are found to be DNase I hypersensitive in adult endothelial cells, whereas only 30% of all other embryonic enhancers are DNase I hypersensitive in endothelia (Figures S2A–S2C). Similarly, 73% of enhancers that are active in embryonic heart tissue are DNase I hypersensitive within cells derived from human heart and great vessel structures, whereas only 27% of all other embryonic enhancers are DNase I hypersensitive in these cell types (Figure S2D).
We also found striking correlation between the number of primitive tissues in which enhancer activity was detected and the number of definitive cell types in which a DHS was detected at that enhancer (Figure 2D). Together, these results suggest both systematic developmental persistence of DNase I hypersensitivity at a subset of early developmental enhancers and a persistent imprint of enhancer functional spectra in the form of DHS patterning across different definitive cell types.
Restriction versus Expansion of the Chromatin Landscape during Differentiation
The relationship between ESCs and definitive lineage derivatives in Figure 1, combined with evidence for developmental persistence of individual DHSs, prompted us to analyze in detail the relative gain and loss of DHSs along specific developmental clines. It is notable that ESCs have the largest DHS complement of all cell types analyzed (n = 257,172 excluding ChrX/Y; Figure S3C and Table S1), of which 58% are shared with at least one definitive cell type (Figure S3D).
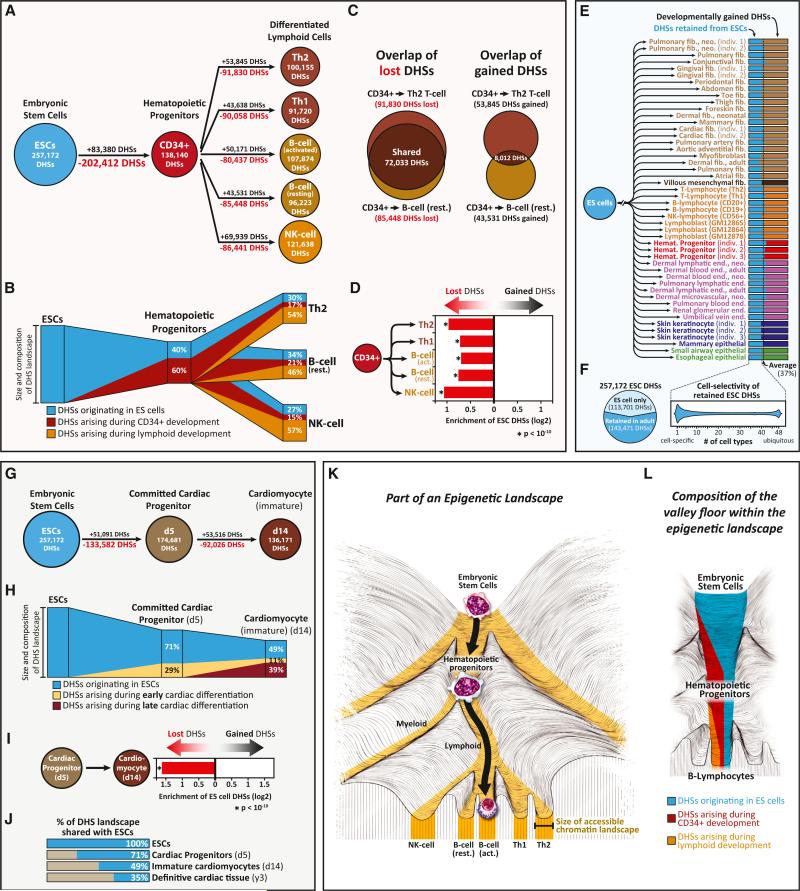
Development along the hematopoietic lineage has been extensively characterized at both the cellular and molecular levels (Orkin, 1995). During hematopoiesis, the accessible chromatin landscape undergoes substantial reorganization that is dominated by the inactivation rather than the de novo activation of DHSs (Figures 3A and 3B). Comparison of the DHS landscape of ESCs to that of hematopoietic progenitors reveals a net loss of 119,032 DHSs, achieved through the silencing of 202,412 ES DHSs and the de novo activation of 83,380 DHSs. As hematopoietic progenitors terminally differentiate into B or T cells, they preferentially inactivate a common set of ~72,000 early developmental DHSs, while activating an average of ~52,000 chiefly lineage-restricted DHSs along each terminal branch (Figures 3C and 3D). Of note, roughly half of the regulatory DNA landscape of each definitive lymphoid cell type is retained from hematopoietic progenitors and roughly one-third from ESCs (Figure 3B).
Figure 3. Developmental Extinction, Maintenance, and De Novo Activation of Chromatin Accessibility at Regulatory DNA.
(A and B) Composition of developing hematopoietic regulatory landscapes. (A) Shown are acquired (black) versus extinguished (red) DHSs during hematopoietic developmental transitions. (B) Schematic illustrating the number of inherited versus acquired DHSs during hematopoietic developmental transitions. The lymphoid DHS compartment colored blue comprises a strict subset of the hematopoietic progenitor DHS compartment colored in blue.
(C and D) Preferential extinction of common early developmental DHSs during development. (C) Comparison of DHSs lost (left) or acquired (right) during the differentiation of hematopoietic progenitors. (D) Shown is the enrichment of ESC DHSs within lost versus gained DHS compartments. p values were calculated using the hypergeometric test.
(E and F) Formulaic composition of terminal regulatory DNA landscapes. (E) Shown is the proportion of the DHS landscape from each definitive cell type that is shared with ESCs. This proportion remains nearly constant at ~37%. (F) Approximately 60% of the ESC DHS landscape persists in varying combinations in definitive cell types (left), and each definitive cell type appears to retain a different cohort of ESC DHSs (right).
(G–J) Regulatory DNA landscape of cardiac differentiation. (G) Acquired (black) versus extinguished (red) DHSs during cardiac differentiation. (H) Schematic illustrating the number of inherited versus gained DHSs during cardiac differentiation. (I) Enrichment of ESC DHSs within lost versus gained DHS compartments. p value was calculated using the hypergeometric test. (J) Clock-like extinction of ESC DHSs during cardiac differentiation.
(K and L) The epigenetic landscape valley floor. (K) Differentiation is accompanied by the progressive restriction of the size of the accessible regulatory DNA landscape (e.g., a narrowing of the epigenetic landscape's valley floors) (gold). (L) Schematic showing restriction, perpetuation, and de novo activation that accompanies differentiation.
See also Figure S3.
Fixed Contribution of ES Regulatory DNA to Terminal Regulatory Landscapes
We next asked how cells differ with respect to the proportion of their regulatory landscapes shared with ESCs. The total number of autosomal DHSs in definitive cell types varied >2-fold, from ~91.7K in Th1 cells to 225.6K in dermal fibroblasts. Surprisingly, within each cell-type landscape, the proportion of DHSs shared with ESCs remained nearly constant, averaging ~37% (Figure 3E). Of these, the vast majority were distal, nonpromoter elements (Figure S3E). In total, across all cell types, approximately 56% of ES DHSs were retained in some definitive cells. (Figure 3F). However, the specific complement of ES DHSs apportioned to each cell type was unique (Figure 3F).
Regulatory Landscape Dynamics during Directed Differentiation from ESCs
To analyze differentiating regulatory DNA landscape dynamics prospectively, we next profiled DHSs during the controlled differentiation of ESCs along the cardiac axis (Yang et al., 2008). During cardiac differentiation under defined conditions, committed cardiac progenitors emerge at day 5 and beating cardiomyocytes at day 14 (Paige et al., 2012). We produced DHS maps for each of these stages (Table S2), as well as from adult heart tissue (as ES-derived cardiomyocytes do not reach full maturity and exhibit primitive features similar to those of early-fetal-stage heart) (Paige et al., 2012).
During directed cardiac differentiation, we observed large-scale reorganization of the DHS landscape (Figure 3G), including inactivation of early developmental elements, extensive forward propagation of ESC and progenitor DHSs, and de novo activation of differentiation-stage-selective elements (Figures 3H and 3I). The inactivation of ESC DHSs during differentiation occurred in a progressive, nearly clock-like fashion, dropping from 71% at day 5, to 49% at day 14, and 35% in adult heart tissue (Figure 3J). Notably, the proportion of ESC DHSs in the terminally differentiated adult cardiac landscape (35%) closely matches the average of other terminally differentiated cells (37%; Figure 3E).
Together, the above findings indicate that the process of lineage differentiation is accompanied by three basic alterations to the regulatory DNA landscape: (1) pruning of ESC DHSs as a function of developmental maturity (Figure 3K); (2) extensive forward propagation of regulatory DNA from progenitors to more defined cells (Figure 3L); and (3) de novo activation of a (generally smaller) number of lineage-restricted DHSs (Figure 3L).
Patterning of Regulatory DNA by Known and Novel Lineage Regulators
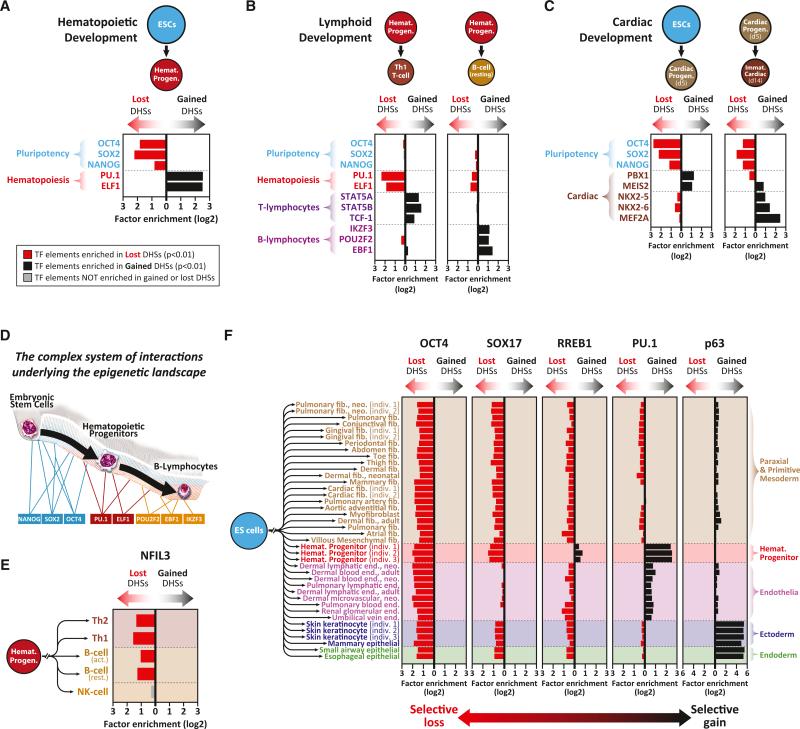
We reasoned that uncovering the TFs that interact with developmentally dynamic (i.e., lost or gained) regulatory DNA should facilitate identification of regulators of cellular identity. During the transition from ESCs to hematopoietic progenitors, lost DHSs were significantly enriched in recognition sequences for pluripotency factors (OCT4, SOX2, NANOG), whereas gained DHSs were enriched in recognition sequences for hematopoietic master regulators including PU.1 and ELF1 (Orkin, 1995) (Figure 4A). By contrast, the subsequent transition from hematopoietic progenitors to T cells, B cells, or NK cells results in selective loss of DHSs enriched in hematopoietic master-regulator recognition sequences and selective gain of DHSs enriched in major T cell, B cell, or NK cell-lineage-regulator recognition sites, respectively (Orkin, 1995) (Figures 4B and S4A). Notably, the presence of TF recognition sequences in DHS peaks is highly predictive of occupancy of the cognate TF, as measured using ChIP-seq or genomic footprinting (Neph et al., 2012c; Samstein et al., 2012). These results indicate that the TFs critical for a given cell state can be identified through analysis of both DHSs lost during transition away from that state and DHSs gained during transition into that state.
Figure 4. Selective Loss versus Gain of DHSs Targeted by Lineage Regulators.
(A–C) Enrichment of lineage regulators in developmentally dynamic DHSs. Enrichment of binding elements for three pluripotency TFs (blue), eight hematopoietic lineage and sublineage TFs (red/purple), and five cardiac lineage TFs (brown) in DHSs lost versus gained during (A) the differentiation of ESCs into hematopoietic progenitors, (B) the differentiation of hematopoietic progenitors into Th1 T cells or B cells, and (C) the differentiation of ESCs into cardiac progenitors and immature cardiomyocytes. p values were calculated using the hypergeometric test.
(D) The “guy-ropes” modeling the epigenetic landscape. Sequential activation of transcriptional regulators underpins the topology and trajectory of the epigenetic landscape—paralleling Waddington's regulatory genes and “guy-ropes” (Waddington, 1957).
(E and F) Lineage-restricted TFs enable the selective retention of regulatory DNA. (E) DHSs containing binding elements for the NK cell master regulator NFIL3 are selectively retained by NK cells and lost in all other lineages. (F) Shown is enrichment of binding elements for five lineage regulators in the DHSs gained versus lost during lineage differentiation from ESCs.
See also Figure S4.
Unlike hematopoietic development, few potent regulators of cardiac differentiation have been characterized. Analysis of the differentiation of ESCs to early cardiac progenitors (d5) revealed selective loss of DHSs enriched in recognition sites for pluripotency factors, coupled with gain of DHSs enriched in motifs for both the well-described early cardiac regulator PBX1 (Chang et al., 2008) and the novel cardiac regulator MEIS2 (Paige et al., 2012) (Figure 4C, left). By contrast, the subsequent transition from cardiac progenitors (d5) to early cardiomyocytes (d14) featured loss of DHSs enriched in PBX1 recognition sites and the appearance of DHSs enriched in binding elements for late cardiac regulators NKX2-5, NKX2-6, and MEF2A (Lyons et al., 1995; Tanaka et al., 2001) (Figure 4C, right).
The above analyses further implicated previously unrecognized lineage-defining roles for numerous other TFs (Figure S4B). For example, the binding landscape for RREB1 contracts in every lineage except for hematopoietic progenitors, where it expands (Figure S4B), indicating that this repressor may play an important yet uncharacterized role in hematopoiesis. Together, these findings indicate that regulatory DNA dynamics during specific developmental transitions reflect the actions of both known and novel lineage-regulating TFs (Figure 4D).
Coordinated Deactivation of Alternative Regulatory Programs during Differentiation
We observed that recognition sites for TFs regulating a given lineage were selectively enriched in the DHSs inactivated along other lineage paths. For example, the recognition landscape for the NK cell master regulator NFIL3 (Gascoyne et al., 2009) remains largely unchanged during NK differentiation but greatly contracts during T cell and B cell development (Figure 4E). Similarly the regulatory landscape for the endothelial regulator SOX17 (Liao et al., 2009) remains largely unchanged during endothelial development but contracts during development of all other lineages (Figure 4F). Similar lineage-restricted patterns were observed for many factors including PU.1, RREB1, and OCT4 (Figures 4F and S4B). This suggests that (1) the regulatory DNA target landscape for certain lineage-restricted factors is largely prepositioned in progenitor cell types via DHSs that contain cognate binding elements and (2) such DHSs are selectively inactivated along lineage paths in which the lineage-relevant TF is lacking. Consequently, development according to a specific lineage program combines the orchestrated activation of lineage-restricted regulatory elements with programmed extinction of regulatory DNA associated with alternative lineage fates (Figure 7D). The latter should in turn serve to passively reinforce lineage commitment.
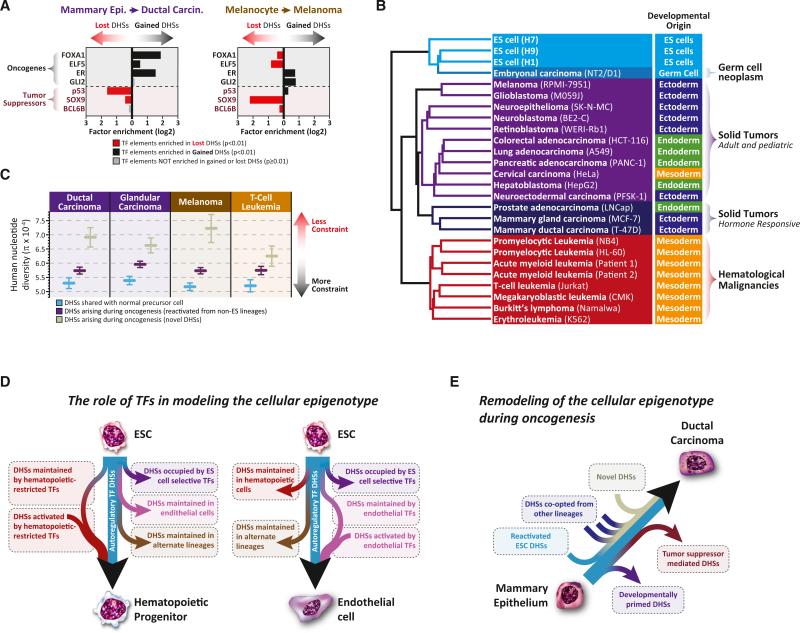
Figure 7. TF Drivers, Functional Organization, and Evolutionary Pressures on Cancer Regulatory Landscapes.
(A) Oncogenic versus tumor-suppressor TF targets in DHSs gained versus lost during oncogenesis. Shown is the enrichment of binding elements for four TF oncogenes and three TF tumor suppressors in the DHSs lost versus gained during the oncogenic transformation of mammary epithelium and melanocytes.
(B) Functional reorientation of the malignant regulatory DNA landscape. Clustering of DHSs from 23 cancer cell types and three different ES lines. Note the predominance of functional or phenotypic features over embryological origins.
(C) DHSs arising during oncogenesis show relaxed evolutionary constraint. Human nucleotide diversity measurements (π, y axis) at DHSs retained from nonmalignant predecessors but not present in ESCs (blue); DHSs arising during oncogenesis that are ectopically activated from alternative lineages, excluding ESCs (purple); and DHSs arising during oncogenesis that are novel to cancer (gray). Error bars represent 95% confidence intervals.
(D and E) Models of normal development and oncogenesis. Shown are the actions of different TF classes on the developing regulatory DNA landscape of hematopoietic progenitors and endothelial cells (D) as well as the oncogenic transformation of mammary epithelium (E).
See also Figure S7.
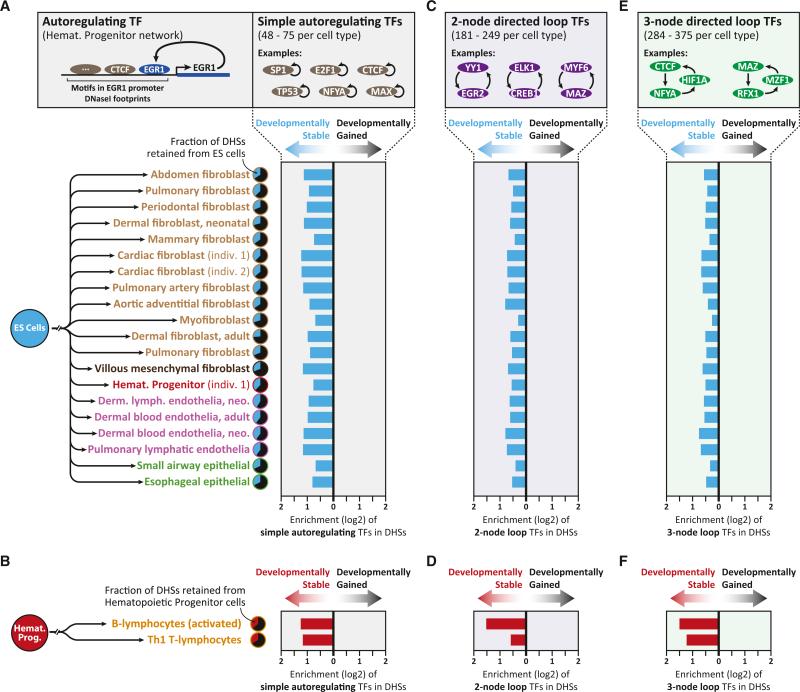
“Memory” DHSs Are Chiefly Occupied by TFs that Regulate Their Own Expression
Many biological processes are perpetuated by reinforcing feedback loops. For example, TFs involved in autoregulatory feedback loops can stabilize their expression during cell division and development (Ptashne et al., 1980; Alon, 2006). We therefore asked whether developmentally stable DHSs were occupied by TFs with autoregulatory features. Using TF-regulatory network maps constructed from genomic footprinting of 23 of the cell types studied here (Neph et al., 2012b), we identified, on average, 68 simple autoregulating TFs per cell type (range 48–75) (Figure 5A). In every cell type analyzed, relative to developmentally dynamic DHSs, developmentally stable DHSs were chiefly and preferentially populated with recognition sites for these simple autoregulating TFs (Figures 5A, 5B, S5A, and S5C), as well as TFs involved in two-node directed and three-node directed loop-network architectures (Figures 5C–5F, S5A, and S5C), which enable indirect autoregulatory behavior. These findings were similar for both developmentally stable distal and promoter-associated DHSs (Figures S5B and S5C).
Figure 5. Epigenetically Stable DHSs Are Potentiated by TFs that Regulate Their Own Expression.
(A) Enrichment of DNase I-footprinted binding elements for simple autoregulating TFs in developmentally stable (i.e., retained from ESCs) versus developmentally gained DHSs in 20 cell types with annotated regulatory networks (Neph et al., 2012b).
(B) Enrichment of DNase I-footprinted binding elements for simple autoregulating TFs in developmentally stable DHSs retained from hematopoietic progenitors, excluding sites retained from ESCs.
(C–F) Enrichment of DNase I-footprinted binding elements for TFs involved in two-node (C and D) or three-node (E and F) autoregulatory loops in developmentally stable versus developmentally gained DHSs. All enrichments shown are significant (p < 10–10, hypergeometric distribution).
See also Figure S5.
Retrograde Remodeling of the Regulatory DNA Landscape during Oncogenesis
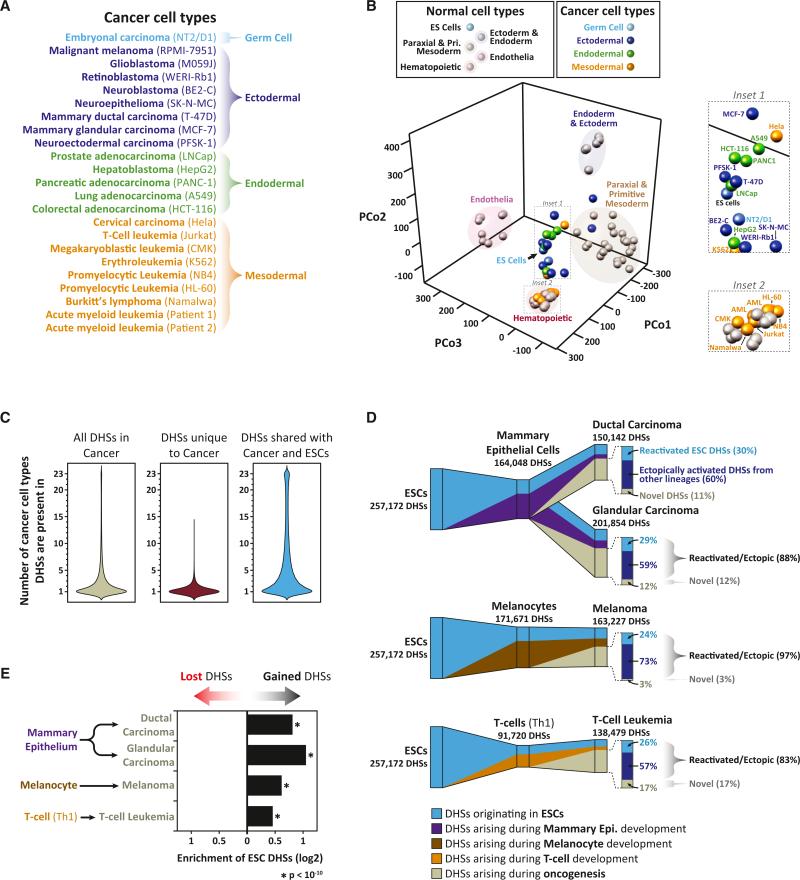
We next explored how the DHS landscape reorganizes during a major pathological deviation. We produced DHS maps from 21 diverse cancer cell lines plus purified sorted cells from two primary malignancies (two subtypes of acute myelogenous leukemia arising in different unrelated individuals) (Figure 6A), yielding between 74,292 and 209,903 autosomal DHSs per cancer cell type (Table S3). We then used PCoA to compare the DHS landscapes of cancer cells with those of normal cells. Whereas the regulatory DNA landscapes of normal cell types are clearly separated (Figures 1C and 6B), cancer DHS landscapes converged on those of ESCs (Figure 6B). Hematological malignancies were a notable exception, forming a distinct group toward the ESC facing pole of the hematopoietic lineage cluster (Figures 6B and S6A).
Figure 6. Retrograde Remodeling of the Regulatory DNA Landscape during Oncogenesis.
(A) Developmental origins of 21 cancer cell lines and 2 primary cancers analyzed with genome-wide DNase I mapping.
(B) PCoA of normal versus malignant cell-type relationships. Shown are normal and cancer cell types projected into a three-dimensional principal coordinates space. Cell-type coloring is indicated above. Note the prominent clustering of cancer cell types around ESCs (inset 1).
(C) Cell selectivity of cancer cell DHSs. Distribution of the number of cancer cell types in which (left) a DHS found in any cancer cell type is observed, (middle) a DHS that is unique to cancer cell types and not found in any of the normal cell types is observed, and (right) a DHS shared between a cancer cell type and ESCs is observed.
(D) Disordered retrograde remodeling of the accessible regulatory DNA landscape during oncogenesis. (D) Shown is the contribution of DHSs found in nonmalignant predecessors to the DHS landscape of each of four cancer cell types. Cancer DHS landscapes are partitioned into DHSs originating in ESCs and maintained in nonmalignant predecessors (blue), DHSs retained from nonmalignant predecessors but not present in ESCs (purple/brown/orange), and DHSs arising during oncogenesis (gray). (Right) Proportion of DHSs arising during oncogenesis that are reactivated (i.e., not inherited) ESC DHSs (light blue), ectopically activated DHSs from alternative lineages (dark blue), or novel DHSs unique to each cancer cell type (gray).
(E) Shown is the enrichmentof ESC DHSs within lost versus gained DHS compartments duringoncogenesis. p values were calculated using the hypergeometric test. See also Figure S6 and Table S3.
To quantify further the apparent retrograde remodeling of cancer regulatory DNA landscapes, we focused our analysis on four malignancies for which DHS maps were available from the presumed corresponding normal precursor cell type (melanocytes for melanoma; mammary epithelium for breast cancer [two types]; and Th1 cells for T cell leukemia). Compared with normal counterparts, all four cancer cell types exhibited substantial reorganization of their DHS landscape in a largely cell-specific manner (Figures 6C, 6D, S6B, and S6C). This reorganization had three major components: (1) reactivation of silenced ESC DHSs (Figure 6E); (2) ectopic activation of DHSs from lineage programs different than that in which the malignancy arose; and (3) appearance of a small proportion of novel DHSs not detected in any normal cell type. Overall, the vast majority (88%–97%) of DHSs activated during oncogenesis were found in some other normal adult or fetal cell or tissue type (Figure 6D). Notably, the regulatory DNA landscape activated during oncogenesis differed substantially between different cancer cell types, with no DHS active in all cancer cell types but no normal cell types (Figures 6C and S6C). Together these results indicate that oncogenesis is characterized by the aberrant co-option of regulatory DNA from ESCs and alternative lineage programs, with each cancer cell type activating a distinct set of elements.
TF Drivers of Cancer Regulatory Landscapes
We next sought to identify TFs mediating reorganization of the regulatory DNA landscape during oncogenesis. The ability of many TFs to function as oncogenes or tumor suppressors is well known (Persson and Leder, 1984). Analysis of DHSs arising in cancers revealed significant enrichment of recognition sites for known oncogenic TFs. By contrast, DHSs lost during oncogenesis were significantly enriched in recognition sequences for known tumor-suppressor TFs. For example, the target landscape of FOXA1 specifically and significantly expands in breast cancer cells compared with normal breast epithelium (Figure 7A), consistent with the role of FOXA1 in mediating estrogen-receptor-dependent chromatin remodeling in breast cancer (Carroll et al., 2005). Furthermore, the target landscape of SOX9 specifically and significantly contracts in melanoma cells compared with normal melanocytes, consistent with the role of SOX9 as a potent melanoma tumor suppressor (Figure 7A) (Passeron et al., 2009). This analysis revealed a variety of TFs with similar patterns of target landscape expansion or contraction in normal versus tumor cells (Figures 7A and S7A), exposing potential novel cell-selective oncogenic and tumor-suppressor roles for such TFs.
Functionally Distorted Memory of Normal Lineage Programs in Malignant Cells
Next we asked whether the transformed regulatory landscapes of cancer cells maintained systematic memory of earlier developmental fate decisions, akin to normal cells (Figure 1). Clustering 23 cancer cell types based on their DHS patterns yielded well-defined clusters (Figure 7B). However, unlike those seen in Figure 1 for normal cells, cancer DHS clusters were typified by functional characteristics of the cancers, rather than their developmental origin For example, hormone-responsive cancers (LNCap, T-47D, and MCF-7) formed a tight cluster, distinct from those of other adult and pediatric solid tumors, germ cell neoplasms, and hematological malignancies. These findings suggest that oncogenic transformation of the regulatory DNA landscape is accompanied by loss of developmental information and can be dominated by selective activation of regulatory elements associated with the derived phenotype of the cancer.
Reduced Evolutionary Pressure on Cancer Regulatory Landscapes
Nucleotide diversity (pi) calculated with genomic sequence data from multiple unrelated individuals provides a quantitative assessment of the extent of ongoing purifying selection at DHSs within the human population (Vernot et al., 2012; Thurman et al., 2012; Neph et al., 2012c). To investigate whether DHSs activated during oncogenesis exhibit levels of selective constraint similar to normal developmentally patterned DHSs, we calculated pi for these two classes of DHSs. Cancer cell DHSs gained during oncogenesis evinced significantly higher nucleotide diversity compared with those retained from normal development (Figures 7C, S7B, and S7C). Of note, similar numbers of DHSs are activated during both normal development and oncogenesis (Figure 6D), yet elements activated during oncogenesis are under less constraint, suggesting that cancer cells selectively recruit regulatory elements that are active at other developmental stages yet may play secondary roles in normal developmental processes. Such reactivation events are likely generated through dysregulation of key developmental TFs (Figure 7A) and can have a large effect on the expression of neighboring genes (Akhtar-Zaidi et al., 2012).
DISCUSSION
The salient findings recounted above can be recapitulated as follows: First, developmental fate and lineage relationships can be derived from the genomic patterning of DHSs in definitive cells; this patterning is distinct from the information conveyed by gene expression. Second, lineage differentiation is associated with three features: (1) extensive propagation of DNase I hypersensitivity at regulatory DNA; (2) pruning of DHSs shared with ESCs as development progresses; and (3) blossoming of a smaller number of lineage-restricted DHSs—all resulting in a more restricted, specialized DHS landscape. Third, pruning of DHSs shared with ESCs during differentiation is proportional to the size of a cell's DHS landscape, with the result that the regulatory DNA landscapes of terminally differentiated cells retain a nearly constant proportion of DHSs shared with ESCs. Fourth, developmentally stable DHSs densely encode binding sites for self-regulating TFs. Finally, in contrast to normal cells, cancer regulatory landscapes feature both extensive reactivation of silenced ESC DHSs and ectopic activation of regulatory DNA from noncognate developmental lineages.
We interpret the above findings to signify a central role for DHS patterning in propagating cellular state and fate information during development and abrogation of this role by oncogenesis. Below we place these findings in historical context and consider both the features of differentiation that Waddington presaged as well as a number of novel and telling insights that our results afford into basic developmental mechanisms and strategies.
From Epigenesis to Epigenetics
The generation of consistent body plans by the sequential differentiation of totipotential material is a foundational conceptual paradigm for development. First articulated by Aristotle (De generatione animalium 739a), this concept was termed “epigenesis” (literally, “moving toward coming into being”) by Harvey (Harvey, 1651) and was widely accepted by the early 20th century (Patten, 1920). Waddington's enduring epigenetic landscape paradigm crystallized this concept and added two important features (Waddington, 1940, 1957). The first concerned the stability of cellular phenotypes, which Waddington depicted as the valley walls within the epigenetic landscape; these valley walls act to guide the cell down a particular “pathway of change that is equilibrated in the sense that the system tends to return to it after disturbance” (Waddington, 1957). The second feature was mechanistic: the proposition that the epigenetic landscape itself is controlled by a complex system of “regulatory genes” that interact with one another in a combinatorial and temporally coordinated fashion to shape the topography of epigenesis (Waddington, 1957).
In view of our results, Waddington's paradigm provides a remarkably prescient schematization of the transformation of the regulatory DNA landscape during development, indicating that global DHS maps provide a missing quantitative dimension for major facets of epigenesis. The fact that proper lineage-branching relationships can be recovered from DHS data but not from gene-expression data suggests that the DHS compartment contains both elements involved in active cellular processes as well as “marker” or “memory” DHSs that preserve information about prior developmental states. Such information is directly evident in persistent DHSs at tissue-selective early developmental enhancers.
Role of TFs in Propagation of Active versus Repressive Chromatin States
Our data indicate that changes in the DHS landscape during development are orchestrated by specific combinations of lineage-restricted TFs (Figure 4) and that developmentally stable DHSs are chiefly occupied by autoregulatory TFs (Figure 5). These results emphasize the central influence of the cellular TF-regulatory network on modeling the regulatory DNA landscape during development (Figures 4D and 7D). It is notable that other examples of “epigenetic memory” chiefly feature propagation of repressive chromatin states through mechanisms such as CpG methylation (Bird, 2002; Kim et al., 2010), histone H3K9 trimethylation (Hathaway et al., 2012), or polycomb (Cavalli and Paro, 1998). Because TFs rapidly reassociate with daughter DNA strands following replication and can bookmark accessible regulatory DNA through mitosis (Egli et al., 2008), TF binding within DHSs is mechanistically well-suited for the propagation of accessible chromatin to daughter cells without invoking other modifications to the chromatin template.
Developmental Transformation of the Regulatory DNA Landscape
Development and differentiation, irrespective of lineage, balance the propagation, extinction, and de novo activation of chromatin accessibility at regulatory DNA in a highly formulaic manner. During development, the regulatory DNA landscape undergoes progressive restriction that outstrips the de novo activation of lineage-restricted DHSs—metaphorically, a narrowing of the epigenetic landscape's valley floors (Figures 3K and 3L). Interestingly, this process appears to be recursive, operating anew from intermediate pluripotential states such as hematopoietic stem cells in a manner reminiscent of classical finite state automata (Turing, 1937). Critically, selective DHS pruning involves the wholesale loss of DHSs associated with alternative fates (Figure 4), thus cementing or “canalizing” a particular developmental pathway in lockstep with differentiation.
Temporal “Memory” of Cellular State and Fate
A remarkable feature of the developmentally stable DHS compartment is its association with cellular maturity. As cells differentiate, ESC-originated DHSs are pruned in a progressive, almost clock-like fashion. Consequently, simply measuring the proportion of DHSs within a cell's regulatory landscape that are shared with ESCs may provide a quantitative measure of developmental maturity.
Relation to the Developmental Hourglass
The concept that developmental biology can provide insights into evolution was sparked by Von Baer's observation that embryos from diverse organisms converge on a common form during the pharyngula stage of mid-embryogenesis (Von Baer, 1828). This point of convergence has been termed the “phylotypic” stage (Cohen, 1963) and coincides with the activation of major developmental regulators such as Hox genes (Duboule, 1994). These observations gave rise to an hourglass model of development (Duboule, 1994; Raff, 1996). Our data in Figure 1D accord with this model and indicate that the hourglass phenomenon may be grounded within discrete sets of regulatory DNA regions.
Connecting Epigenesis and Oncogenesis
Oncogenesis is accompanied by the drastic remodeling of the DHS landscape, resulting in a loss of developmental information, and reversion to a “pseudoprimitive” state that combines regulatory DNA features of ESCs with those of other developing lineages (Figures 6 and 7). This state is not truly _de_-differentiated—which implies walking back along a path previously taken—but rather _dys_-differentiated, having aberrantly co-opted “normal” regulatory elements from alternative lineage paths (Figure 7E). As such, cancer cells encompass a multidimensional deviation from normal development and can no longer be placed on Waddington's landscape. This feature may explain the long-standing observation that oncogenesis is accompanied by the reappearance of fetal antigens (Hirszfeld et al., 1932; Tatarinov, 1964). Notably, this finding is difficult to reconcile with models of oncogenesis that posit cancer origins from developmental remnants. If cancer cells simply arose from uncontrolled proliferation of a more primitive cell remnant, we would expect cancer cells to retain strong lineage signatures; however, with the exception of hematological malignancies, this is decidedly not the case.
The process of _dys_-differentiation appears to result from the misregulation of key developmental TFs (Figure 7A). However, despite the growth and phenotypic advantages bestowed by the transformed regulatory DNA landscape, malignant cells have likely lost many of the beneficial regulatory redundancies and feedback mechanisms that are formed during normal development and that maintain epigenetic stability in the face of environmental and genetic stress (Figures 7D and 7E). It is tempting to speculate that this patchwork reorganization of the chromatin landscape during oncogenesis may expose exploitable vulnerabilities of the malignant state.
EXPERIMENTAL PROCEDURES
DHS Mapping
All cell types were subjected to nuclear isolation, DNase I digestion, DNase I double-hit fragment purification and library construction, and high-throughput sequencing, as described previously (Thurman et al., 2012; John et al., 2011; Hesselberth et al., 2009). Data from additional cell types were utilized from Thurman et al. (2012). DHSs were computed for each cell type at an FDR of 1% as previously described (John et al., 2011).
DHS Clustering
We used the BEDOPS suite to generate a reference multiset union of DHSs across all cell types (Neph et al., 2012a). The Euclidean distance between two cell types was calculated using binary peak calls for each DHS within the reference set for a given cell type. Pairwise Euclidean distances between all cell types were clustered using the nearest-neighbor algorithm. For cancer cell types, we utilized Euclidean distances and Ward clustering.
Comparison of Adult versus Embryonic Enhancer Activity
Data for tests of human enhancers in a mouse developmental model (Pennacchio et al., 2006) were downloaded from http://enhancer.lbl.gov/.
Autoregulating TFs
Comprehensive maps of human TF-regulatory networks constructed with genome-wide DNase I footprint maps were used to identify TFs that form autoregulatory loops (Neph et al., 2012b). The occupancy of these autoregulatory TFs within developmentally stable and developmentally gained DHSs was mapped using TF-binding elements contained within DNase I footprints (Neph et al., 2012c).
Human Nucleotide Diversity Measurements
Human nucleotide diversity measurements (p) were calculated using whole-genome sequences from 53 unrelated individuals as previously described (Vernot et al., 2012).
Supplementary Material
01
ACKNOWLEDGMENTS
This work was supported by NIH grants U54HG004592, U54HG007010, and U01ES017156 to J.A.S.; NIH grant P30DK056465 to S.H.; and NIH grants P01GM081719, U01HL100405, P01HL094374, and R01HL084642 to C.E.M. We thank many colleagues, particularly Joseph Costello (UCSF), for contributing cells for DNase I mapping and expression profiling. A.B.S. was supported by grant FDK095678A from NIDDK. S.L.P. was supported by NHLBI grant F30HL095343. We thank Rae Senarighi (UW) for expert assistance with graphic design. All data from this study are available through the ENCODE data repository at UCSC (http://www.encodeproject.org) and the Roadmap Epigenomics data repository at NCBI (http://www.ncbi.nlm.nih.gov/epigenomics). All data from this study are free to use and are not subject to consortium embargo dates.
Footnotes
REFERENCES
- Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336:736–739. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. First Edition Chapman and Hall/CRC; London: 2006. [Google Scholar]
- Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard J. Embryological aspects and etiology of carcinoma. Lancet. 1902;159:1758–1761. [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Chang C-P, Stankunas K, Shang C, Kao S-C, Twu KY, Cleary ML. Pbx1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development. 2008;135:3577–3586. doi: 10.1242/dev.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Living Embryos: An Introduction to the Study of Animal Development. Pergamon Press; New York: 1963. [Google Scholar]
- Domazet-Lošo T, Tautz D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468:815–818. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- Duboule D. Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Dev. Suppl. 1994;1994:135–142. [PubMed] [Google Scholar]
- Egli D, Birkhoff G, Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat. Rev. Mol. Cell Biol. 2008;9:505–516. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJM. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- Gleason DF, Mellinger GT. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J. Urol. 1974;111:58–64. doi: 10.1016/s0022-5347(17)59889-4. [DOI] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Groudine M, Weintraub H. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell. 1982;30:131–139. doi: 10.1016/0092-8674(82)90019-8. [DOI] [PubMed] [Google Scholar]
- Harvey W. Exercitationes de Generatione Animalium. Typis Du-Gardianis; Impensis O. Pulleyn; London: 1651. [Google Scholar]
- Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirszfeld L, Halber W, Rosenblat J. Untersuchungen uber Verwandtschaftsreaktionen zwischen embryonal und Krebsgewebe. II. Menschenembryo und Menschenkrebs. ZeitschriftfUr Immunitatsforschung und Experimentelle Therapie. 1932;75:209–216. [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung M-HH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, Ohler U, Bergman CM, Tomancak P. Gene expression divergence recapitulates the developmental hourglass model. Nature. 2010;468:811–814. doi: 10.1038/nature09634. [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LIR, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao WP, Uetzmann L, Burtscher I, Lickert H. Generation of a mouse line expressing Sox17-driven Cre recombinase with specific activity in arteries. Genesis. 2009;47:476–483. doi: 10.1002/dvg.20520. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Regeneration in the egg, embryo, and adult. Am. Nat. 1901;35:949–973. [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, et al. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012a;28:1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012b;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012c;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH. Transcription factors and hematopoietic development. J. Biol. Chem. 1995;270:4955–4958. doi: 10.1074/jbc.270.10.4955. [DOI] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sand-strom R, Pabon L, Reinecke H, Pratt G, Keller G, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passeron T, Valencia JC, Namiki T, Vieira WD, Passeron H, Miyamura Y, Hearing VJ. Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. J. Clin. Invest. 2009;119:954–963. doi: 10.1172/JCI34015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten B. The Early Embryology of the Chick. P. Blakiston's Son & Co. Inc.; Philadelphia: 1920. [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Persson H, Leder P. Nuclear localization and DNA binding properties of a protein expressed by human c-myc oncogene. Science. 1984;225:718–721. doi: 10.1126/science.6463648. [DOI] [PubMed] [Google Scholar]
- Ptashne M. On the use of the word ‘epigenetic’. Curr. Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Jeffrey A, Johnson AD, Maurer R, Meyer BJ, Pabo CO, Roberts TM, Sauer RT. How the lambda repressor and cro work. Cell. 1980;19:1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- Pujadas E, Feinberg AP. Regulated noise in the epigenetic landscape of development and disease. Cell. 2012;148:1123–1131. doi: 10.1016/j.cell.2012.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff RA. The Shape of Life: Genes, Development, and the Evolution of Animal Form. University of Chicago Press; Chicago, IL: 1996. [Google Scholar]
- Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sand-strom R, Neph S, Sabo P, Kim JM, Liao W, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Schinke M, Liao HS, Yamasaki N, Izumo S. Nkx2.5 and Nkx2.6, homologs of Drosophila tinman, are required for development of the pharynx. Mol. Cell. Biol. 2001;21:4391–4398. doi: 10.1128/MCB.21.13.4391-4398.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarinov IuS. Detection of embryo-specific alpha-globulin in the blood serum of a patient with primary liver cancer. Vopr. Med. Khim. 1964;10:90–91. [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turing AM. On computable numbers, with an application to the entscheidungs problem. Proc. Lond. Math. Soc. 1937;42:230–265. [Google Scholar]
- Vernot B, Stergachis AB, Maurano MT, Vierstra J, Neph S, Thurman RE, Stamatoyannopoulos JA, Akey JM. Personal and population genomics of human regulatory variation. Genome Res. 2012;22:1689–1697. doi: 10.1101/gr.134890.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Baer KE. Über Entwicklungsgeschichte der Thiere. Beobachtung und Reflexion. Gebrüder Bornträger; Königsberg: 1828. [Google Scholar]
- Waddington CH. Cancer and the theory of organiser. Nature. 1935;135:606–608. [Google Scholar]
- Waddington CH. An Introduction to Modern Genetics. The Macmillan Company; New York: 1939. [Google Scholar]
- Waddington CH. Organisers and Genes. Cambridge University Press; Cambridge, UK: 1940. [Google Scholar]
- Waddington CH. The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. George Allen & Unwin; Sydney: 1957. [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01