Starving our Microbial Self: The Deleterious Consequences of a Diet Deficient in Microbiota-Accessible Carbohydrates (original) (raw)
. Author manuscript; available in PMC: 2016 Jun 7.
Published in final edited form as: Cell Metab. 2014 Aug 21;20(5):779–786. doi: 10.1016/j.cmet.2014.07.003
Abstract
The gut microbiota of a healthy person may not be equivalent to a healthy microbiota. It is possible that the Western microbiota is actually dysbiotic and predisposes individuals to a variety of diseases. The asymmetric plasticity between the relatively stable human genome and the more malleable gut microbiome suggests that incompatibilities between the two could rapidly arise. The Western lifestyle, which includes a diet low in microbiota-accessible carbohydrates (MACs), has selected for a microbiota with altered membership and functionality compared to those of groups living traditional lifestyles. Interactions between resident microbes and host leading to immune dysregulation may explain several diseases that share inflammation as a common basis. The low-MAC Western diet results in poor production of gut microbiota-generated short-chain fatty acids (SCFAs), which attenuate inflammation through a variety of mechanisms in mouse models. Studies focused on modern and traditional societies, combined with animal models, are needed to characterize the connection between diet, microbiota composition, and function. Differentiating between an optimal microbiota, one that increases disease risk, and one that is causative or potentiates disease will be required to further understand both the etiology and possible treatments for health problems related to microbiota dysbiosis.
The Gut Microbiota: A Control Center for Human Biology
The human gut harbors a diverse ecosystem of trillions of microbial cells (Costello et al., 2012; Qin et al., 2010). The traditional view that gut microbiota effects are limited to the host's digestive tract, or more recently extended to metabolism and immune status, has given way to the realization that these microbes can have widespread impact on diverse aspects of a host's physiology (Sommer and Backhed, 2013). Externally applied forces, such as diet or antibiotics, result in rapid alterations of the microbiota, thereby affecting the status of host-microbiota relations (David et al., 2014; Dethlefsen et al., 2008; Dethlefsen and Relman, 2010; Walker et al., 2011). The malleability of our gut residents is further reframing this community's role as a platform for rational manipulation of host biology: the microbiota provides a manipulatable lever to improve human health and to treat or prevent disease (Sonnenburg and Fischbach, 2011; van Nood et al., 2013; Vrieze et al., 2012).
At a more fundamental level, the very plasticity that makes the gut microbiota an attractive therapeutic avenue can result in unintentional and maladaptive changes, or dysbiosis. A plethora of microbiota enumeration studies in recent years have implicated microbiota dysbiosis in a growing list of Western diseases, such as metabolic syndrome, inflammatory bowel disease, and cancer. These studies rely on comparing the microbiota of individuals that are diseased to those that are healthy. The contribution of the dysbiotic microbiota to disease phenotype has been illustrated via microbiota transplant in several instances (Koren et al., 2012; Liou et al., 2012; Ridaura et al., 2013; Turnbaugh et al., 2006; Vijay-Kumar et al., 2010). However, use of the term “dysbiotic” must be accompanied by the recognition that the definition of a healthy microbiota that should serve as a frame of reference is still poorly defined. It is possible that the microbiota of a healthy Westerner is also dysbiotic, significantly increasing the risk of developing diseases characterized by excessive or inappropriate immune inflammatory responses.
Microbiota-Accessible Carbohydrates: Food for the Masses
Much of the carbon and energy for members of the microbiota originate from plant- and animal-derived dietary carbohydrates (Koropatkin et al., 2012; Salyers et al., 1977). These carbohydrates are composed of monosaccharides that are connected through numerous types of glycosidic linkages and, in some instances, further modified by chemical substituents like acetyl and sulfate groups. The variation in their chemical composition, solubility, and size differentiates these carbohydrates into a vast array of ecological niches. However, microbial competition within the gut is intense for metabolic access to the energy and carbon sequestered in these molecules. Residents of the microbiota are equipped with a specialized collection of enzymes such as glycoside hydrolases and polysaccharide lyases, which can break down complex carbohydrate linkages into consumable oligosaccharides or monosaccharides (Cantarel et al., 2009; Martens et al., 2011). A model microbiota containing ~160 species has in excess of 9,000 glycoside hydrolases and 2,000 polysaccharide lyases, which serve as the gateway to a complex microbial food web within the gut (El Kaoutari et al., 2013). When extrapolating to ~1,000 species, a single human gut microbiota may contain upward of 60,000 carbohydrate-degrading enzymes. The human genome, on the other hand, has only a small number of glycoside hydrolases (~17) and no polysaccharide lyases that are involved in carbohydrate digestion within the gut.
Carbohydrates for gut microbe fermentation can come from a variety of sources including (1) dietary or host-derived animal glycans (2) synthesized by other microbes that are food-borne (e.g., the yeast cell wall) or a gut resident, and (3) dietary plant material, commonly referred to as dietary fiber, which is the most common fuel for the microbiota of many humans (Flint et al., 2012; Koropatkin et al., 2012). The role of dietary fiber on human health has been discussed for decades. In the 1960s and 1970s, Denis Burkitt and Hugh Trowell documented the significantly larger intake of dietary fiber by Africans relative to Westerners and their coincident lack of Western diseases such as diabetes, heart disease, and colorectal cancer. Many of these diseases, including asthma, allergies, and inflammatory bowel diseases, appear in young Westerners and are therefore not simply a product of our modern extension of lifespan, but share a dysregulated immune system as a common basis. Burkitt reported that rural Africans passed stool that was up to five times greater by mass, had intestinal transit times that were more than twice as fast, and ate three to seven times more dietary fiber (60–140 g versus 20 g) than their Western counterparts (Burkitt et al., 1972). However, the mechanisms by which dietary fiber would exert positive health effects were unknown. Burkitt and others hypothesized that the increased transit time and stool size that accompanied a high-fiber diet may serve to dilute potentially hazardous microbial metabolic products and quickly remove them from the colonic lining (Burkitt and Trowell, 1977; Trowell and Burkitt, 1986). But recent studies are suggesting that the missing mechanistic explanation for the beneficial effects of dietary fiber may be largely attributed to fermentation by the microbiota.
In discussing carbohydrate substrates that fuel the colonic microbial ecosystem, dietary fiber is a problematic term commonly employed for lack of a better option. The definition of dietary fiber has evolved over the past 60 years since the concept was first introduced (Hipsley, 1953; Trowell, 1976; Trowell et al., 1976; Trowell and Burkitt, 1987). Current use of this term is complicated by multiple definitions associated with different official organizations (Raninen et al., 2011). In addition, multiple laboratory tests may be used to determine “dietary fiber” content specified on nutritional labels, none of which provides an accurate measure of what the definitions specify. The most standard method of quantifying dietary fiber content neglects many types of carbohydrates that are destined for microbiota fermentation in the colon, like inulin, but includes noncarbohydrate entities like lignin. In addition, the term dietary fiber encompasses the carbohydrates that are fermentable by an individual's microbiota plus those that remain unfermented and serve a bulking role (Kashyap et al., 2013a). Attempts to use measurements of soluble verses insoluble fiber as approximations of fermentable versus nonfermentable, respectively, have proven inaccurate. Similar to “dietary fiber,” the term “plant polysaccharides” is a common euphemism for “microbiota food,” but this category excludes important nonplant carbohydrates and oligosaccharides.
When referring to the carbohydrates that can be metabolically used by gut microbes, we propose the term “microbiota-accessible carbohydrate” (MAC; see Box 1 for expanded discussion of MACs). MACs serve as selective agents, altering the composition of the microbiota, but also dictate the functionality and metabolic output. Multiple studies have shown that diet can serve as a selective potent force in reshaping the microbiota (David et al., 2014; Walker et al., 2011; Wu et al., 2011). Highly controlled experiments in mice have shown that the quantity and type of carbohydrates that feed the microbiota alter simplified and complex microbial communities (Faith et al., 2011; Kashyap et al., 2013a; Sonnenburg et al., 2010; Turnbaugh et al., 2009b). Humans eating controlled diets supplemented with nonstarch polysaccharides or resistant starch, both with high levels of potential MACs, exhibit diet-induced alterations in microbiota composition (Walker et al., 2011). Due to the “gateway” role of primary fermenters within the colonic ecosystem, MACs can promote certain microbes either directly (those that consume a substrate) or indirectly (via crossfeeding interactions) (Fischbach and Sonnenburg, 2011). Despite a large gap in our understanding of how the diversity and quantity of dietary MACs influence host health, many studies are strengthening the links between MACs, microbiota diversity and metabolism, and host health.
MACs and Microbiota Diversity
Accumulating data suggest that host health is linked to dietary MAC-induced alterations in microbiota composition and diversity. A dietary intervention performed on 49 obese individuals showed an increase in microbiota gene richness when participants reduced energy intake and increased dietary fiber consumption for 6 weeks (Cotillard et al., 2013). In mice, reduction of dietary fiber has a direct effect on microbiota diversity and short-chain fatty acid production; the latter is an indication that indeed the dietary fiber includes MACs (Kashyap et al., 2013b; Trompette et al., 2014). Recently, a significant and reversible difference in microbiota composition was observed in humans that shifted to either a diet of all plants or one of all animal products, illustrating the impact diet has on microbiota composition (David et al., 2014). However, despite significant increases in dietary fiber (and presumably MAC) intake over the 4-day plant-based diet, an increase in microbiota diversity was not observed. Four days of increased dietary MACs may be an insufficient time period to for previously rare (undetectable) species to flourish or for environmentally acquired microbes to entrench. It remains to be determined whether the simplified Western microbiota has lost species that cannot be recovered upon increased dietary MACs.
Additional human studies have correlated increased dietary MAC consumption to microbiota composition, diversity, and health. The gut microbiota of children from a rural village in Burkina Faso revealed stark differences compared to a “healthy” Western cohort in Italy (De Filippo et al., 2010). In addition to disparity in represented phylotypes, relative to the Europeans, the African group showed (1) an altered ratio of the two dominant phyla of the human microbiota, with increased representation of Bacteroidetes relative to Firmicutes (the opposite of the Bacteroidetes depletion associated with obesity in several studies) (Ley et al., 2005, 2006; Turnbaugh et al., 2009a); (2) increased microbiota diversity; and (3) large increases of SCFA production including 4-fold more butyrate and propionate. These changes coincide with nearly twice the dietary fiber intake in the African group compared to the European group.
Our Ancient Microbiota
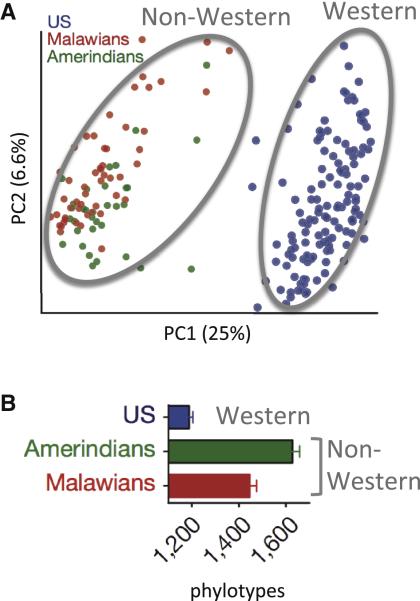
While the Burkina Faso study strengthened the link between diet and microbiota diversity, it also provided the first glimpse at what the human microbiota may have looked like during the Neolithic revolution in agriculture as humans embraced cultivated foods. An independent study of rural groups in Malawi and Venezuela similarly revealed greater bacterial diversity and stark differences in microbiota composition of nonmodern societies relative to Western groups (Figure 2). The microbiota from all three traditional agrarian groups in Malawi, Venezuela, and Burkina Faso were more similar to one another in microbiota diversity and SCFA production than to their Western counterparts (Yatsunenko et al., 2012). In addition to supporting the view that the modern microbiota deviates substantially from ancestral states, these findings point to the need to infer the microbiota at additional time points of human evolution, such as that of preagricultural humans.
Figure 2. The Western Microbiota Diverges from That of Non-Western Populations.
(A) Principle coordinate plot of gut microbiota composition in individuals (each dot represents one person's microbiota) from three populations based on Unifrac distances of the bacterial 16S rRNA profile. Distance between dots represents extent of compositional difference.
(B) Average number of bacterial phylotypes detected within the gut microbiota from three populations; error bars indicate SEM. Data reprinted with permission (Yatsunenko et al., 2012).
Hunting and gathering was the lifestyle that dominated for the longest time span in our species existence. The Hadza, a group of ~1,000 people in Tanzania, are the last full-time hunter-gatherers in Africa and represent one of the best modern approximations of preagricultural humans (Marlowe, 2010). A recent study comparing the microbiota of 27 Hadza to 16 Italians, who represent a Western cohort, showed many interesting differences, including a relative enrichment of the Prevotella genus, the presence of Treponema species, and the lack of Bifidobacterium (Schnorr et al., 2014). Perhaps their most significant finding is the much larger extent of bacterial diversity in the foraging microbiota compared to the industrialized group.
Interpretation of these studies comes with important caveats. Most studies focus on the bacterial component of the gut microbiota, excluding important archaea, eukaryotes, and viruses. All of these rural populations are composed of modern people that approximate, but are not exact representations of, a human ancestral state. Furthermore, it is almost certain that there was no single “ancestral state” for any given time in human evolution. As humans radiated across the planet, diverse lifestyles, diets, locations, and genetic differences likely resulted in microbiota variation within populations; it is certain that even gradients of transculturation exist within each indigenous cohort. So additional microbiota data, coupled with metadata about diet and lifestyle from these and other traditional populations, will be required to place these initial findings into context. Paleogenomics offer an alternative avenue to elucidate, rather than infer, microbiotas of the past (Tito et al., 2012). The major dietary shifts occurring between the hunter-gatherer lifestyle, early Neolithic farming, and more recently during the Industrial Revolution are reflected in changes in microbial membership within dental tartar of European skeletons throughout these periods (Adler et al., 2013). Notably, the transition to our modern lifestyle has been accompanied by the appearance of cariogenic bacteria and markedly reduced oral microbiota diversity.
Microbiota Metabolism and Host Health
Two independent studies of individuals in Europe have illustrated that microbiome diversity is linked to human health (Cotillard et al., 2013; Le Chatelier et al., 2013). The 341 combined participants from both studies were comprised of obese, overweight, and healthy individuals and exhibited a bimodal distribution of microbiota gene content: some individuals exhibited low gene count (LGC) and others high gene count (HGC). The LGC group was more insulin resistant and had higher fasting triglycerides, higher LDL cholesterol, and higher markers of inflammation relative to the HGC group. Dietary intervention of 49 obese or overweight individuals (18 from the LGC group and 27 from the HGC group) resulted in loss of fat mass and was accompanied by improvements in clinical markers such as lipid and insulin values, insulin resistance, and measures of inflammation for both the LGC and HGC groups (Cotillard et al., 2013). However, improvements in these clinical markers were most pronounced for the HGC group, indicating that microbiome richness was a predictor of response to diet-induced improvements. Dietary intervention did result in an increase in gene richness in the LGC cohort that approached but still remained significantly lower than the HGC. Together these independent studies indicate that metabolic disease clinical phenotypes are associated with gene content diversity of the gut microbiota and that diet can directly impact this diversity.
The mechanisms underlying the link between microbiota diversity and health are still unknown, but SCFAs appear to be one likely mediator. It is possible that microbiota diversity is an intermediate correlate of high-MAC diets and that fermentation end-products absorbed into circulation are important effectors of host physiology. The short-chain fatty acids, acetate, propionate, and butyrate, are common end-products of carbohydrate-fermenting microbes in the distal gut (Wong et al., 2006), and several factors, including dietary substrates, can affect the relative ratios and concentrations (Macfarlane and Macfarlane, 2003; Topping and Clifton, 2001). SCFAs are absorbed by the host and serve as calories salvaged from otherwise inaccessible carbohydrates, but they also play diverse regulatory roles including regulating histone acetylation and signaling through G protein coupled receptors (Bergman, 1990; Brown et al., 2003; Davie, 2003). Recently several groups have linked microbiota-generated SCFAs to anti-inflammatory effects. SCFAs increase the pool of regulatory T cells in the gut, and MAC-dependent SCFAs produced by the gut microbiota are protective of allergic airway inflammation in a mouse model (Arpaia et al., 2013; Furusawa et al., 2013; Smith et al., 2013; Trompette et al., 2014). Multiple studies indicate a dependence upon fatty-acid, G protein-coupled receptors (GPR41 or GPR43).
SCFAs regulate energy homeostasis, a role to which the beneficial metabolic effects of fiber have been attributed (Layden et al., 2013). Butyrate appears to broadly influence metabolism and circulating propionate signals via the afferent nervous system to influence gluconeogenesis in the intestine, as well as results in improved metabolic profiles and reduced weight gain (De Vadder et al., 2014; Gao et al., 2009; Lin et al., 2012). Additional studies have linked dietary fiber and the resulting SCFAs to protection from diet-induced obesity (Cani et al., 2007; Lin et al., 2012; Neyrinck et al., 2012; Zhou et al., 2009). It remains to be determined how dietary MAC consumption influences the thousands of microbiota-produced metabolites and the resulting influence on host biology (Marcobal et al., 2013; Nicholson et al., 2012; Russell et al., 2011, 2013).
Effects from decreased SCFA output on the low-MAC Western diet may be compounded by changes in microbiota localization and barrier disruption in the distal gut. The paucity of fermentable carbohydrates selects for a community of microbes that are adept at eating host-derived MACs found in the intestinal mucus layer. Consumption of host mucins by many gut species serves as a consistent endogenous source of energy for the microbiota in the absence of dietary polysaccharides (Sonnenburg et al., 2005). The long-term effects of low dietary MACs on the gut mucus depletion, barrier function, and inflammation have not been studied in detail; however, genetic mucus depletion in mice results in microbiota-induced inflammation and ultimately colitis (Johansson et al., 2014; Van der Sluis et al., 2006).
The Future: Looking to the Past
Whether the ancient microbiota is a better microbiota is debatable; however, at least two arguments support this idea: (1) our human genome lacks the plasticity that our microbiota possesses and therefore has not been able to keep pace with the recent profound changes brought on by our modern lifestyle, such as the consequences of a diet very low in microbiota-accessible carbohydrates (e.g., low diversity microbiota, less SCFA production); and (2) traditional societies typically have much lower rates of Western diseases (Wirsing, 1985).
The plasticity of our microbiota in response to diet was likely highly adaptive in an ancient environment. In delegating part of our digestion and calorie harvest to our gut residents, the microbial part of our biology could easily adjust to day-to-day or season-to-season variation in available food. Considering the remarkable resilience of the gut community to short-term perturbation, it is likely that microbiota adaptation is largely reversible on short time scales (David et al., 2014; Dethlefsen and Relman, 2010; Lee et al., 2013). However, the ability of the microbiota to accommodate dietary change may be deleterious to its own maintenance and ultimately to our health in the context of MAC deprivation that has occurred over generations in industrialized countries. In addition to the loss of microbial species with which we have coevolved, the lack of MACs required to fuel this community results in profound functional changes, such as decreases in SCFA production.
Undoubtedly, sudden changes in human diet throughout evolution resulting from major lifestyle changes (e.g., cooking, hunting with weapons, and farming) were accompanied by fundamental shifts in microbiota composition and function (Figure 3A). These new configurations of the microbiota likely introduced suboptimal interactions with many aspects of human biology; however, such interactions would be expected to optimize over millennia. It is possible that the new microbiota configuration resulting from modern diet and lifestyle presents incompatibilities with our human biology, the ill effects of which contribute significantly to many modern diseases (Figure 3B) (Blaser and Falkow, 2009).
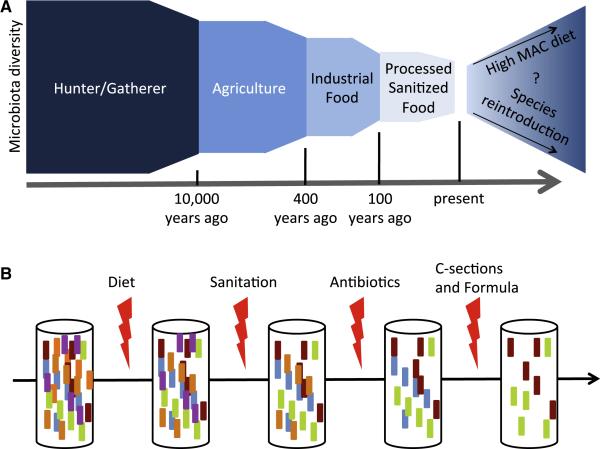
Figure 3. The Multiple-Hit Hypothesis for How the Microbiota of Industrialized Societies Has Lost Diversity over Time.
(A) Microbiota diversity was likely altered at multiple stages of human evolution. As diversity and quantity of dietary MACs decreased with agriculture, industrialized food production, and processed food, the model reflects data that indicate a corresponding decrease in microbiota diversity.
(B) While diet is likely a key mediator of microbiota diversity, additional technological and medical leaps, while providing solutions for important problems such as infectious disease, have likely served as insults to the microbiota. These multiple hits have prevented the maintenance of microbiota diversity over recent generations.
Significant questions remain unanswered to address how large of a role the Western microbiota plays in modern diseases. How important is the loss of coevolved microbial species versus the loss of microbial metabolites? Would a more sensible diet that increases MAC content be sufficient to recover microbiota diversity, or have species been irreversibly lost? Does imposing a high-MAC diet on a Western microbiota present new incompatibilities and potential unanticipated negative health consequences? We must be aware that ongoing dissemination of the industrialized lifestyle and processed foods to the most remote areas of the globe will be accompanied by a series of microbial extinction events similar to that which has left the biodiversity of the Western microbiota decimated. While imposing poorer health on populations as they modernize, the global eradication of critical microbiota members will likely have far-reaching importance. These remaining untouched populations may represent valuable repositories of important taxa that have been lost from Western populations. Collection and preservation of these microbes followed by their deliberate reintroduction supported by a diet rich in the MACs that sustain them might be the best hope of rewilding the Western microbiota.
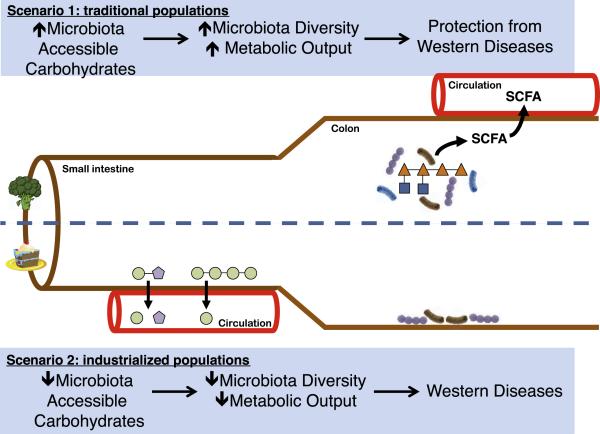
Figure 1. The Divergent Metabolic Scenarios of a High-MAC versus a Low-MAC Diet.
Two scenarios represent a trade-off in how calories are absorbed by the host. In the first scenario, a high-MAC diet that has few simple sugars, the major contribution of carbohydrates to host metabolism is in the form of the SCFA fermentation end-products of the microbiota. In addition to calories, these molecules play diverse regulatory roles in human physiology, including protection from many Western diseases. In the second scenario, the low-MAC Western diet results not only in a loss of beneficial microbial metabolites such as SCFA, but also in selection of a distinct microbiota that may seem foreign to the host. Increased representation of mucus-utilizing microbes, decreased gut motility, and increased calories in the form of sugar and fat may synergize to cause Western diseases. MACs include carbohydrates from diet, host secretion (e.g., mucin glycans), or other resident microbes that serve as a metabolic input for members of the microbiota.
Box 1. Microbiota-Accessible Carbohydrates.
MACs are carbohydrates that are metabolically available to gut microbes. MACs include carbohydrates that are dietary and resistant to degradation and absorption by the host, and they may be secreted by the host in the intestine (e.g., mucus) or produced by microbes within the intestine. Dietary MACs may come from a variety of sources including plants, animal tissue, or food-borne microbes, but must be metabolized by the microbiota. Much of the cellulose humans consume is not metabolized by gut microbes and does not qualify as a MAC (Chassard et al., 2010). The amount of dietary MACs present in a single food source differs for each individual, since which carbohydrates are metabolized depends upon the membership of each person's microbiota. The individuality of which carbohydrates qualify as MACs is illustrated by the presence of genes for the consumption of the algal polysaccharide porphyran in the microbiomes of Japanese individuals, rarely found in North American and European individuals (Hehemann et al., 2010, 2012). For those harboring a porphyran-degrading strain, porphyran would be a MAC; but for those without a microbiota adaptation to seaweed, porphyran would not be a MAC. Similarly, germ-free mice that lack a microbiota might consume a diet high in potential MACs, but none of the carbohydrates would be considered bona fide MACs, since they would transit the digestive tract unmetabolized by microbes. Alternatively, lack of dietary MACs results in a microbial community reliant upon endogenous host-derived MACs in the form of mucin glycans (Sonnenburg et al., 2005). Different host genotypes can influence the identity of MACs available to the microbiota in multiple ways. For example, host genotype can dictate alteration of mucus structures, such as the absence of alpha-1-2 fucose residues in the mucus of nonsecretor individuals that lack alpha-1-2-fucosyltransferase activity in the intestine (Kashyap et al., 2013b). Similarly, host genotype can determine how efficiently carbohydrates are digested and absorbed in the small intestine. For example, lactose becomes a substrate for the microbiota in people who are lactose intolerant, and therefore should be considered a MAC for these individuals. For nursing infants, dietary MACs in breast milk are known as human milk oligosaccharides (Bode, 2012; Marcobal et al., 2010, 2011). Therefore, the term “microbiota-accessible carbohydrate” contributes to a conceptual framework for investigating and discussing the amount of metabolic activity that a specific food or carbohydrate can be expected to produce within a given microbiota (Figure 1).
ACKNOWLEDGMENTS
We thank members of the Sonnenburg lab and Jeff Leach for valuable discussions and Sara Fisher for editing this manuscript. This research was supported by R01-DK085025 and DP2-OD006515. J.L.S. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
REFERENCES
- Adler CJ, Dobney K, Weyrich LS, Kaidonis J, Walker AW, Haak W, Bradshaw CJ, Townsend G, Soltysiak A, Alt KW, et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat. Genet. 2013;45:450–455. doi: 10.1038/ng.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat. Rev. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22:1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- Burkitt DP, Trowell HC. Dietary fibre and western diseases. Ir. Med. J. 1977;70:272–277. [PubMed] [Google Scholar]
- Burkitt DP, Walker AR, Painter NS. Effect of dietary fibre on stools and the transit-times, and its role in the causation of disease. Lancet. 1972;2:1408–1412. doi: 10.1016/s0140-6736(72)92974-1. [DOI] [PubMed] [Google Scholar]
- Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, Gibson GR, Delzenne NM. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassard C, Delmas E, Robert C, Bernalier-Donadille A. The cellulose-degrading microbial community of the human gut varies according to the presence or absence of methanogens. FEMS Microbiol. Ecol. 2010;74:205–213. doi: 10.1111/j.1574-6941.2010.00941.x. [DOI] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, et al. ANR MicroObes Consortium Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie JR. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003;133(Suppl):2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA. 2010;108(Suppl. 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, Cefalu WT, Ye J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- Hehemann JH, Kelly AG, Pudlo NA, Martens EC, Boraston AB. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc. Natl. Acad. Sci. USA. 2012;109:19786–19791. doi: 10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipsley EH. Dietary “fibre” and pregnancy toxaemia. BMJ. 1953;2:420–422. doi: 10.1136/bmj.2.4833.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjövall H, Hansson GC. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, Sonnenburg ED, Ferreyra JA, Higginbottom SK, Million M, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013a;144:967–977. doi: 10.1053/j.gastro.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc. Natl. Acad. Sci. USA. 2013b;110:17059–17064. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Bäckhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat. Rev. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layden BT, Angueira AR, Brodsky M, Durai V, Lowe WL., Jr. Short chain fatty acids and their receptors: new metabolic targets. Transl. Res. 2013;161:131–140. doi: 10.1016/j.trsl.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. MetaHIT Consortium Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Lin HV, Frassetto A, Kowalik EJ, Jr., Nawrocki AR, Lu MM, Kosinski JR, Hubert JA, Szeto D, Yao X, Forrest G, Marsh DJ. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE. 2012;7:e35240. doi: 10.1371/journal.pone.0035240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou AP, Paziuk M, Luevano JM, Jr., Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci. Trans. Med. 2012;5:178ra141. doi: 10.1126/scitranslmed.3005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, Mills DA. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 2010;58:5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, Spor-mann A, Fischbach MA, Sonnenburg JL. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlowe F. The Hadza: Hunter-Gatherers of Tanzania. University of California Press; Berkeley: 2010. [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyrinck AM, Possemiers S, Verstraete W, De Backer F, Cani PD, Delzenne NM. Dietary modulation of clostridial cluster XIVa gut bacteria (Roseburia spp.) by chitin-glucan fiber improves host metabolic alterations induced by high-fat diet in mice. J. Nutr. Biochem. 2012;23:51–59. doi: 10.1016/j.jnutbio.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raninen K, Lappi J, Mykkänen H, Poutanen K. Dietary fiber type reflects physiological functionality: comparison of grain fiber, inulin, and polydextrose. Nutr. Rev. 2011;69:9–21. doi: 10.1111/j.1753-4887.2010.00358.x. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WR, Gratz SW, Duncan SH, Holtrop G, Ince J, Scobbie L, Duncan G, Johnstone AM, Lobley GE, Wallace RJ, et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 2011;93:1062–1072. doi: 10.3945/ajcn.110.002188. [DOI] [PubMed] [Google Scholar]
- Russell WR, Hoyles L, Flint HJ, Dumas ME. Colonic bacterial metabolites and human health. Curr. Opin. Microbiol. 2013;16:246–254. doi: 10.1016/j.mib.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Salyers AA, West SE, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl. Environ. Microbiol. 1977;34:529–533. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Comm. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Backhed F. The gut microbiota—masters of host development and physiology. Nat. Rev. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Fischbach MA. Community health care: therapeutic opportunities in the human microbiome. Sci. Transl. Med. 2011;3:78ps12. doi: 10.1126/scitranslmed.3001626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tito RY, Knights D, Metcalf J, Obregon-Tito AJ, Cleeland L, Najar F, Roe B, Reinhard K, Sobolik K, Belknap S, et al. Insights from characterizing extinct human gut microbiomes. PLoS ONE. 2012;7:e51146. doi: 10.1371/journal.pone.0051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- Trowell H. Definition of dietary fiber and hypotheses that it is a protective factor in certain diseases. Am. J. Clin. Nutr. 1976;29:417–427. doi: 10.1093/ajcn/29.4.417. [DOI] [PubMed] [Google Scholar]
- Trowell H, Burkitt D. Physiological role of dietary fiber: a ten-year review. Bol. Asoc. Med. P. R. 1986;78:541–544. [PubMed] [Google Scholar]
- Trowell HC, Burkitt DP. The development of the concept of dietary fibre. Mol. Aspects Med. 1987;9:7–15. doi: 10.1016/0098-2997(87)90013-6. [DOI] [PubMed] [Google Scholar]
- Trowell H, Southgate DA, Wolever TM, Leeds AR, Gassull MA, Jenkins DJ. Letter: dietary fibre redefined. Lancet. 1976;1:967. doi: 10.1016/s0140-6736(76)92750-1. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009a;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009b;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Büller HA, Dekker J, Van Seuningen I, Renes IB, Einerhand AW. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsing R. The health of traditional societies and the effects of acculturation. Curr. Anthropol. 1985;26:303–322. [Google Scholar]
- Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Martin RJ, Tulley RT, Raggio AM, Shen L, Lissy E, McCutcheon K, Keenan MJ. Failure to ferment dietary resistant starch in specific mouse models of obesity results in no body fat loss. J. Agric. Food Chem. 2009;57:8844–8851. doi: 10.1021/jf901548e. [DOI] [PMC free article] [PubMed] [Google Scholar]