VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases (original) (raw)
Abstract
The ECS (Elongin B/C-Cul2/Cul5-SOCS-box protein) complex is a member of a family of ubiquitin ligases that share a Cullin-Rbx module. SOCS-box proteins recruit substrates to the ECS complex and are linked to Cullin-Rbx via Elongin B/C. VHL has been implicated as a SOCS-box protein, but lacks a C-terminal sequence (downstream of the BC box) of the SOCS box. We now show that VHL specifically interacts with endogenous Cul2-Rbx1 in mammalian cells, whereas SOCS-box proteins associate with Cul5-Rbx2. We also identify LRR-1 and FEM1B as proteins that share a region of homology with VHL (the VHL box, including the BC box and downstream residues) and associate with Cul2-Rbx1. ECS complexes can thus be classified into two distinct protein assemblies, that is, those that contain a subunit with a VHL box (composed of the BC box and a downstream Cul2 box) that interacts with Cul2-Rbx1, and those that contain a subunit with a SOCS box (BC box and downstream Cul5 box) that interacts with Cul5-Rbx2. Domain-swapping analyses showed that the specificity of interaction of VHL-box and SOCS-box proteins with Cullin-Rbx modules is determined by the Cul2 and Cul5 boxes, respectively. Finally, RNAi-mediated knockdown of the Cul2-Rbx1 inhibited the VHL-mediated degradation of HIF-2α, whereas knockdown of Cul5-Rbx2 did not affect it. These data suggest that the functions of the Cul2-Rbx1 and Cul5-Rbx2 modules are distinct.
Keywords: E3 ubiquitin ligase, VHL-box protein, SOCS-box protein, Cullin
The Elongin B/C complex, a heterodimer composed of ubiquitin-like Elongin B and Skp1-like Elongin C, was initially identified as a positive regulator of the RNA polymerase II elongation factor Elongin A, which is one of several transcription factors capable of increasing the rate of elongation by RNA polymerase II in vitro (Bradsher et al. 1993; Aso et al. 1995; Reines et al. 1999). Elongin B/C was subsequently found to be a component of the von Hippel-Lindau (VHL) tumor suppressor complex (Duan et al. 1995; Kibel et al. 1995). Interaction of Elongin B/C with Elongin A and with the VHL protein depends on the binding of Elongin C to a 10-amino acid degenerate sequence motif, XLXXXCXXX(A,I,L,V), referred to as the BC box (Duan et al. 1995; Kibel et al. 1995; Aso et al. 1996). Analysis of the crystal structure of the VHL-Elongin B/C complex revealed that binding of Elongin C to the BC box of VHL is mediated by interaction of the highly conserved leucine at position 2 of the BC box with a hydrophobic pocket formed by residues in the C-terminal half of Elongin C (Stebbins et al. 1999). Elongin B binds to a short N-terminal region of Elongin C and does not appear to interact directly with the BC box. In addition to its interaction with Elongin A and VHL, Elongin B/C binds to SOCS-box proteins via the N-terminal regions of the SOCS box, whose amino acid sequences are compatible with the consensus sequence of the BC box (Kamura et al. 1998; Zhang et al. 1999). SOCS proteins were initially identified as negative regulators of signal transduction and now include eight isoforms that all share an N-terminal Src homology 2 (SH2) domain and a SOCS box, an ∼40-amino acid motif, in the C-terminal region (Endo et al. 1997; Naka et al. 1997; Starr et al. 1997). A large number of SOCS-box proteins, which contain various motifs, including GTPase, WD40 repeat, ankyrin repeat, and SPRY domains in addition to a SOCS box, has also been identified (Masuhara et al. 1997; Hilton et al. 1998). SOCS-box proteins, together with other proteins with a BC-box motif, including Elongin A and VHL, are thus also referred to as BC-box proteins.
Ubiquitylation and subsequent proteasomal degradation of regulatory proteins control a variety of cellular processes, including cell cycle progression, gene transcription, and signal transduction. The multistage processing of target proteins begins with enzymatic tagging with a polyubiquitin chain and culminates with ubiquitin-dependent degradation by the 26S proteasome (Hochstrasser 1996; Hershko and Ciechanover 1998). Ubiquitin ligases (E3s) are responsible for the recognition and recruitment of target proteins for polyubiquitylation, and Cullin-dependent ubiquitin ligases (CDLs) constitute one large class of E3s (Guardavaccaro and Pagano 2004). These multisubunit E3s contain two core components, a Cullin subunit and the RING finger-containing protein Rbx (also referred to as Roc or Hrt), the latter of which stabilizes the interaction of Cullin with a ubiquitin-conjugating enzyme (E2) (Kamura et al. 1999; Ohta et al. 1999; Seol et al. 1999; Skowyra et al. 1999; Tan et al. 1999). The SCF (Skp1-Cul1-F-box protein) and ECS (Elongin B/C-Cul2/5-SOCS-box protein) complexes are representative CDLs (Bai et al. 1996; Feldman et al. 1997; Skowyra et al. 1997; Kamura et al. 2001). In SCF complexes, F-box proteins are linked to the Cul1-Rbx1 module via the Skp1 adapter protein, which binds to an ∼40-amino acid degenerate sequence motif (the F box) present in F-box proteins. In ECS complexes, Elongin B/C functions as an adapter that links SOCS-box proteins to the Cullin-Rbx module.
VHL forms a multiprotein complex with Elongin B/C, Cul2, and Rbx1 (Kamura et al. 1999), and this complex catalyzes the ubiquitylation of hypoxia-inducible factor-α (HIF-α) after hydroxylation of a critical proline residue within the oxygen-dependent degradation domain of this protein (Maxwell et al. 1999; Cockman et al. 2000; Kamura et al. 2000; Ohh et al. 2000; Tanimoto et al. 2000; Ivan et al. 2001; Jaakkola et al. 2001). Similarly, SOCS1 was shown to interact with Elongin B/C and Cul2 and to target JAK2, Vav, IRS1, and IRS2 for ubiquitylation and proteasomal degradation (De Sepulveda et al. 2000; Kamizono et al. 2001; Rui et al. 2002). On the other hand, the SOCS-box proteins MUF1, WSB1, and SOCS1 were found to associate preferentially with the Cul5-Rbx1 module rather than with Cul2-Rbx1 (Kamura et al. 2001). SOCS-box proteins and VHL have thus been thought to function as the receptor subunits of ECS complexes.
We now show that VHL and SOCS-box proteins specifically bind to endogenous Cul2-Rbx1 and Cul5-Rbx2, respectively, in mammalian cells. This specificity is determined by short sequences (designated Cul2 and Cul5 boxes) that are located C-terminal to the BC box. In addition to VHL, we have also now identified two BC-box proteins, LRR-1 and FEM1B, as Cul2-binding proteins that contain a Cul2 box. The SOCS-box proteins SOCS1, SOCS3, SSB1, SSB2, SSB4, RAR3, WSB1, and MUF1 contain a Cul5 box. The VHL box (BC box plus Cul2 box) and SOCS box (BC box plus Cul5 box) thus appear to define two distinct families of proteins. Our findings suggest that the Cul2 box and the Cul5 box are the key determinants of the association between BC-box proteins and the Cullin-Rbx module, and that Rbx1 and Rbx2 in higher eukaryotes are functionally distinct, at least in terms of their specific binding to Cullin proteins.
Results
VHL-box and SOCS-box proteins bind to endogenous Cul2-Rbx1 and Cul5-Rbx2 modules, respectively
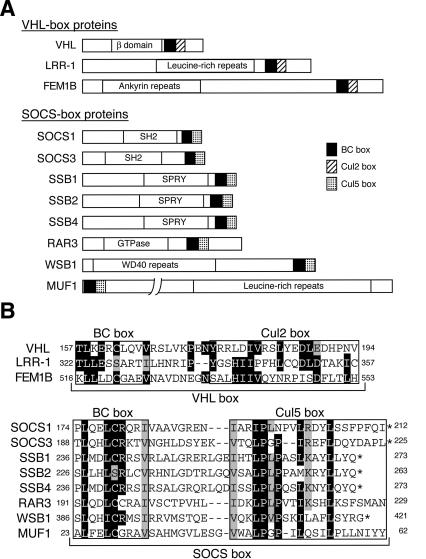
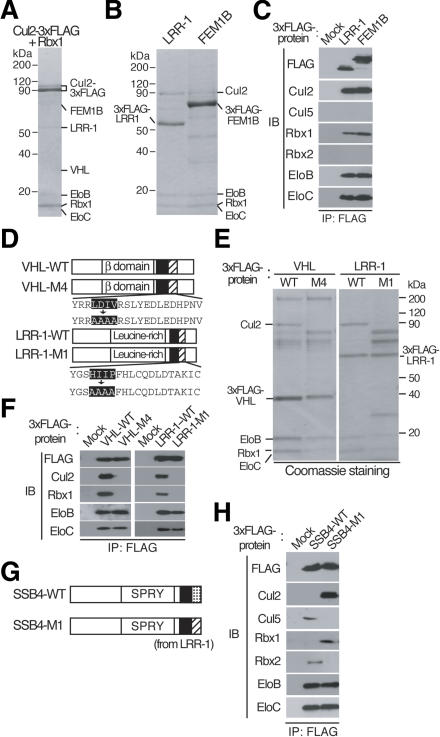
We previously showed that VHL interacts specifically with the Cul2-Rbx1 module under physiological conditions (Kamura et al. 1999), but that it also associates with Cul5-Rbx1 when VHL, Elongin B, Elongin C, Cul5, and Rbx1 are overexpressed in Sf21 insect cells (Kamura et al. 2001). Similarly, in mammalian or insect cells overexpressing all components of the ECS complex, SOCS-box proteins interact with either Cul2-Rbx1 or Cul5-Rbx1 (Kamizono et al. 2001; Kamura et al. 2001). In the present study, we also identified LRR-1 and FEM1B as Cul2-interacting proteins (see Fig. 6, below) that contain a BC box (Fig. 1A). Alignment of the amino acid sequences of 11 proteins that contain the BC box (VHL, LRR-1, FEM1B, SOCS1, SOCS3, SSB1, SSB2, SSB4, RAR3, WSB1, MUF1) revealed that these proteins can be separated into two groups (VHL, LRR-1, and FEM1B in one group, and the remaining eight proteins in the other) on the basis of the sequence downstream of the BC box (Fig. 1B). As demonstrated below, these two distinct sequences appear to be important for the specificity of binding to Cul2 or to Cul5, respectively, and we have therefore designated them the Cul2 box and the Cul5 box. We hereafter refer to the regions containing the BC box and either the Cul2 box or the Cul5 box as the VHL box and the SOCS box, respectively.
Figure 6.
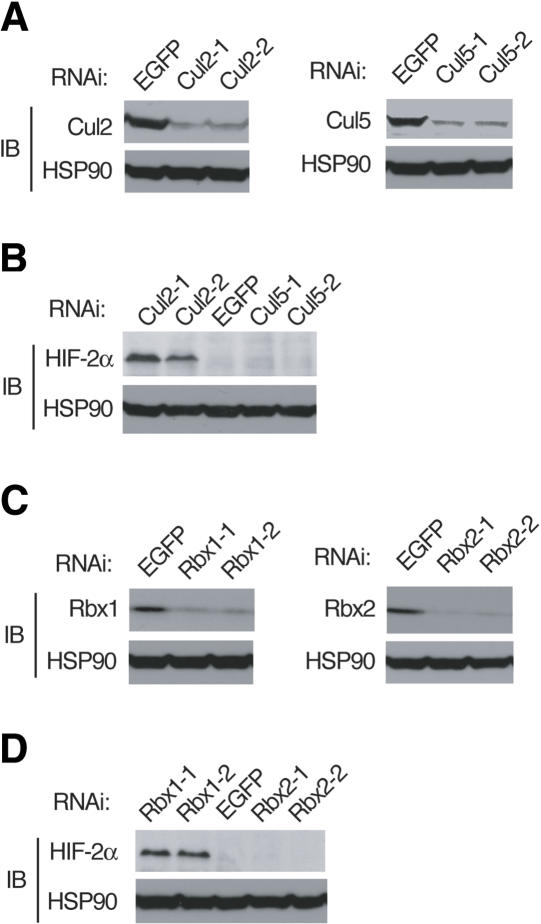
Effect of RNAi knockdown of cullin-Rbx modules on the HIF-2α expression. (A,B) HeLa cells were infected with with a retroviral vector encoding either Cul2 (Cul2-1 or Cul2-2), Cul5 (Cul5-1 or Cul5-2), or EGFP (control) shRNAs. Lysates of the cells were subjected to immunoblot analysis with anti-Cul2, anti-Cul5, anti-HIF-2α, or anti-HSP90 (control). (C,D) Knockdown experiments for Rbx1 (Rbx1-1 or Rbx1-2) or Rbx2 (Rbx2-1 or Rbx2-2) were performed as in A and B.
Figure 1.
Sequence-based classification of VHL-box and SOCS-box proteins. (A) Domain organization of VHL-box and SOCS-box proteins. (B) Alignment of amino acid sequences of VHL-box and SOCS-box proteins. The BC box, Cul2 box, and Cul5 box are indicated (boxed). Identical (black) and similar (gray) amino acids are shaded. Asterisks indicate C termini. Sequences are of the human proteins with the exception of SOCS1, WSB1, and MUF1.
We generated HEK293T cells stably transfected with expression vectors encoding VHL or one of six SOCS-box proteins, all tagged with three copies of the Flag epitope (3xFlag) at their N termini. Proteins that interacted with VHL or the SOCS-box proteins were purified by coimmunoprecipitation with antibodies to (anti-) the Flag epitope, fractionated by SDS-polyacrylamide gel electrophoresis (PAGE), and stained with Coomassie brilliant blue (Fig. 2A). Mass spectrometric analysis of bands corresponding to coprecipitated proteins identified endogenous Elongin B, Elongin C, Cul2, and Rbx1 as associated with 3xFlag-VHL. We failed to detect Cul5 or Rbx2 in these immunoprecipitates in multiple experiments. In contrast, endogenous Elongin B, Elongin C, Cul5, and Rbx2 were identified in the immunoprecipitates prepared from cells expressing 3xFlag-tagged SOCS3, SSB1, SSB2, RAR3, WSB1, or MUF1. Neither Cul2 nor Rbx1 was detected in these precipitates, however. Consistent with the results of mass spectrometry, immunoblot analysis revealed the presence of endogenous Elongin B, Elongin C, Cul2, and Rbx1 (but not Cul5 or Rbx2) in the VHL immunoprecipitates, and the presence of endogenous Elongin B, Elongin C, Cul5, and Rbx2 (but not Cul2 or Rbx1) in the SOCS-box protein immunoprecipitates (Fig. 2B). Characterization of other proteins associated with VHL or the SOCS-box proteins will be described elsewhere (T. Kamura and K.I. Nakayama, in prep.).
Figure 2.
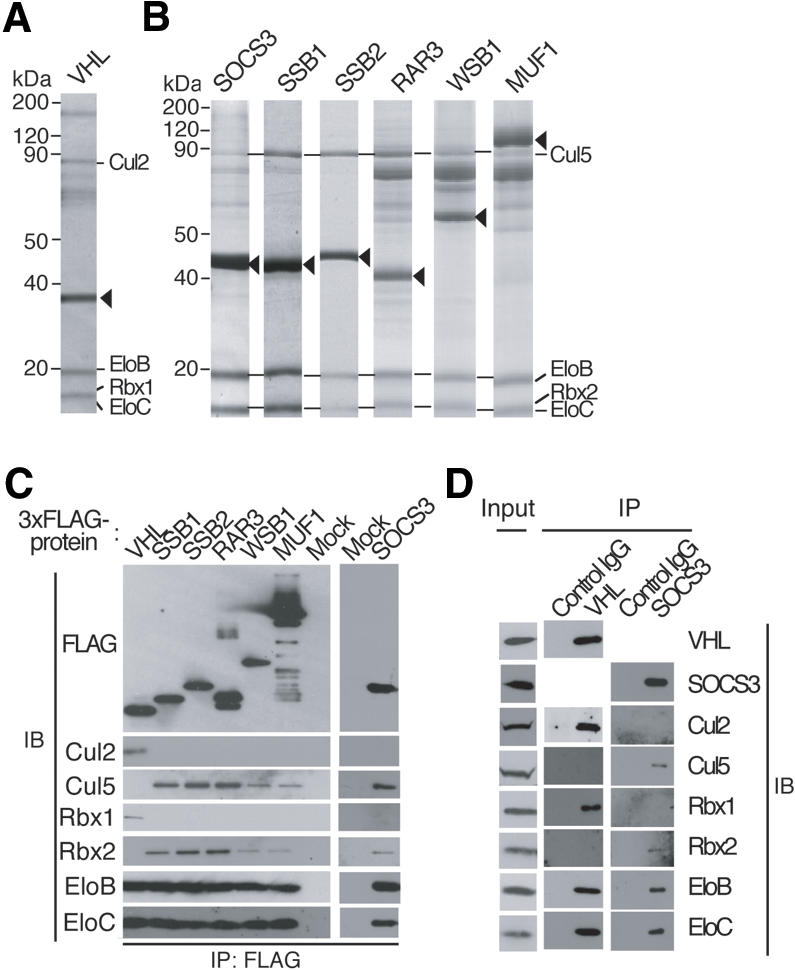
Identification of proteins that associate with VHL or SOCS-box proteins. (A,B) Lysates of HEK293T cells stably expressing the 3xFlag-tagged recombinant proteins indicated at the top of each lane were subjected to immunoprecipitation with anti-Flag, and the resulting precipitates were fractionated by SDS-PAGE and stained with Coomassie blue. Proteins identified by mass spectrometric analysis are indicated, as are the 3xFlag-tagged proteins (arrowheads). (EloB) Elongin B; (EloC) Elongin C. (C) Immunoprecipitates (IP) similar to those obtained in A were subjected to immunoblot analysis (IB) with antibodies to Flag or to the indicated proteins. Mock indicates cells transfected with empty vector. (D) RAW264.7 cells were incubated for 5 h with lipopolysaccharide (10 ng/mL-1) and then for 5 h with 10 μM MG132, after which cell lysates were subjected to immunoprecipitation with either anti-VHL, anti-SOCS3, or control IgG. The cell extracts (1% of the input for immunoprecipitation) and the resulting precipitates were subjected to immunoblot analysis with antibodies to the indicated proteins.
To exclude the possibility that the observed interactions between VHL and the endogenous Cul2-Rbx1 or between SOCS-box proteins and the endogenous Cul5-Rbx2 module were due to overexpression of these proteins, we examined whether endogenous VHL and SOCS3 associate physically with endogenous Cul2-Rbx1 and Cul5-Rbx2, respectively, in mouse monocytic RAW264.7 cells. The cells were cultured with lipopolysaccharide to induce SOCS3 expression and then with the proteasome inhibitor MG132 to facilitate the potential interaction of SOCS3 with Cul5-Rbx2. VHL, Cul2, Cul5, Rbx1, Rbx2, Elongin B, and Elongin C were expressed in this cell line (Fig. 2D). Cell lysates were then subjected to immunoprecipitation with anti-VHL, anti-SOCS3, or control immunoglobulin G (IgG). Immunoblot analysis of the resulting precipitates revealed that endogenous VHL interacted with endogenous Elongin B, Elongin C, Cul2, and Rbx1, but not with Cul5 or Rbx2, whereas SOCS3 interacted with endogenous Elongin B, Elongin C, Cul5, and Rbx2, but not with Cul2 or Rbx1, suggesting that VHL and SOCS3 bind physiologically to the Cul2-Rbx1 and Cul5-Rbx2 modules, respectively.
The Cul5 box is essential for SOCS-box protein association with Cul5-Rbx2
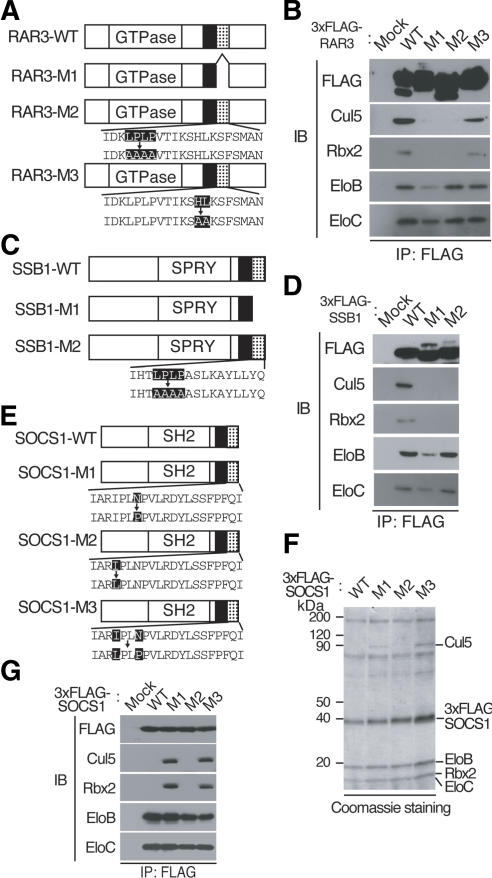
Comparison of the amino acid sequence of VHL and those of SOCS-box proteins suggested that the C-terminal portion (Cul5 box) of the SOCS box might be responsible for the specific interaction of SOCS-box proteins with the Cul5-Rbx2 module (Fig. 1B). To test this hypothesis, we constructed three RAR3 mutants (Fig. 3A) as follows: M1, which contains an internal deletion of residues 209-229 (the Cul5 box); M2, which contains mutations at the conserved positions 212-215 (LPLP → AAAA); and M3, which contains mutations at positions 221 and 222 (HL → AA), the latter of which is also conserved in the Cul5 box. HEK293T cells were transfected with 3xFlag-tagged wild-type RAR3 or the RAR3 mutants M1, M2, or M3, and the abilities of these proteins to interact with endogenous Cul5-Rbx2 were assessed by coimmunoprecipitation and immunoblot experiments. Endogenous Elongin B, Elongin C, Cul5, and Rbx2 were detected in the anti-Flag precipitates obtained from cells expressing wild-type RAR3 or the M3 mutant, whereas only Elongin B and Elongin C were detected in the precipitates obtained from cells expressing the M1 or M2 mutants (Fig. 3B). These data thus suggest that the LPLP sequence in the Cul5 box of RAR3 is required for binding to the Cul5-Rbx2 module. Endogenous Cul2 and Rbx1 were not coimmunoprecipitated with any of the RAR3 derivatives (data not shown).
Figure 3.
The Cul5 box of SOCS-box proteins is required for interaction with the Cul5-Rbx2 module. (A) Domain organization of human wild-type RAR3 (RAR3-WT) and the M1, M2, and M3 mutants thereof. BC-box (filled) and Cul5-box (dotted) are shown. (B) Lysates of HEK293T cells stably expressing the indicated 3xFlag-tagged RAR3 derivatives were subjected to immunoprecipitation with anti-Flag, and the resulting precipitates were subjected to immunoblot analysis with antibodies to the indicated proteins. (C) Domain organization of human wild-type SSB1 (SSB1-WT) and the M1 and M2 mutants thereof. (D) Lysates of HEK293T cells stably expressing the indicated 3xFlag-tagged SSB1 derivatives were subjected to immunoprecipitation with anti-Flag, and the resulting precipitates were subjected to immunoblot analysis with antibodies to the indicated proteins. (E) Domain organization of mouse wild-type SOCS1 (SOCS1-WT) and the M1, M2, and M3 mutants thereof. (F,G) Lysates of HEK293T cells stably expressing the indicated 3xFlag-tagged SOCS1 derivatives were subjected to immunoprecipitation with anti-Flag, and the resulting precipitates were analyzed by SDS-PAGE and mass spectrometry (F) or by immunoblotting (G) as described in Figure 2.
To test the generality of these results, we similarly deleted or mutated the Cul5 box of the SOCS-box protein SSB1 (Fig. 3C) and tested the abilities of the mutant proteins to interact with endogenous Cul5-Rbx2. The M1 mutant, which lacks the Cul5 box (amino acids 257-273), interacted with Elongin B and Elongin C, but not with Cul5 or Rbx2 (Fig. 3D). The M2 mutant, which contains mutations at positions 260-263 (LPLP → AAAA), also failed to bind to the Cul5-Rbx2 module, suggesting that the interaction between SSB1 and Cul5-Rbx2 is mediated by the Cul5 box of SSB1. The Cul5 box, especially the conserved amino acid sequence LPXP, thus appears to be indispensable for the binding of SOCS-box proteins to the Cul5-Rbx2 module.
SOCS1 does not bind to Cul5-Rbx2 because of an incompletely conserved Cul5 box
SOCS1 has been shown to interact with Cul2 and to catalyze the ubiquitylation of JAK2, Vav, IRS1, and IRS2 when these proteins are overexpressed (De Sepulveda et al. 2000; Kamizono et al. 2001; Rui et al. 2002). We examined whether SOCS1 associates with the Cullin-Rbx module in transfected HEK293T cells. Unexpectedly, both mass spectrometric and immunoblot analyses of 3xFlag-SOCS1 immunoprecipitates revealed the presence of Elongin B/C, but not of Cullin-Rbx (Fig. 3E-G). Given that the conserved amino acid sequence LPXP in the Cul5 box is replaced with IPLN in SOCS1, we tested whether mutating this sequence in SOCS1 might affect the ability to bind to Cul5-Rbx2 (Fig. 3E). The M1 and M3 mutants, in which the amino acid sequence IPLN was changed to IPLP and LPLP, respectively, interacted with Cul5-Rbx2, as revealed by mass spectrometric (Fig. 3F) and immunoblot (Fig. 3G) analyses. In contrast, the M2 mutant, in which the sequence IPLN was changed to LPLN, did not bind to Cul5-Rbx2. These results thus provide further support for the importance of the conserved amino acid sequence LPXP in the Cul5 box, especially of proline in the fourth position of the motif, for the association of SOCS-box proteins with the Cul5-Rbx2 module.
VHL chimeric proteins containing a Cul5 box associate with Cul5-Rbx2
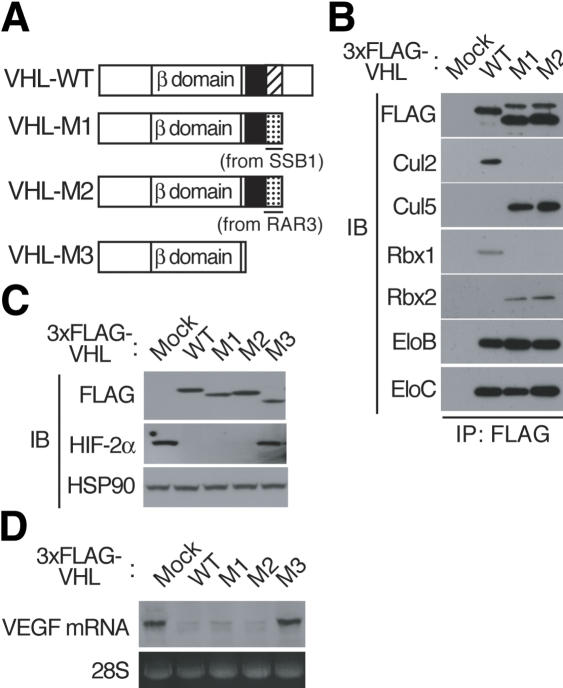
To test further whether the Cul5 box determines specificity in binding to the Cul5-Rbx2 module, we examined the effect of replacing the Cul2 box of VHL with a Cul5 box on the ability to bind to the Cullin-Rbx module. We constructed two such chimeric proteins, VHL-M1 and -M2, in which the N-terminal region of VHL (residues 1-174) containing the β domain and the BC box was fused to the Cul5 box derived from SSB1 (residues 257-273) or from RAR3 (residues 209-229), respectively (Fig. 4A). Vectors encoding 3xFlag-tagged VHL derivatives were introduced into human 786O cells, in which both alleles of the VHL gene are mutated. Coimmunoprecipitation analysis revealed that wild-type VHL associated with Elongin B, Elongin C, Cul2, and Rbx1, whereas the chimeric proteins VHL-M1 and VHL-M2 interacted with Elongin B/C and Cul5-Rbx2 (Fig. 4B). These results thus indicate that the Cul5 box shared by members of the SOCS-box protein family determines the specificity of binding to the Cul5-Rbx2 module.
Figure 4.
Chimeric proteins composed of VHL and a Cul5 box assemble with the Cul5-Rbx2 module. (A) Domain organization of human wild-type VHL (VHL-WT) and the M1, M2, and M3 derivatives thereof. BC-box (filled), Cul2-box (hatched), and Cul5-box (dotted) are shown. (B) Lysates of 786O cells stably expressing the indicated 3xFlag-tagged VHL derivatives were subjected to immunoprecipitation with anti-Flag, and the resulting precipitates were subjected to immunoblot analysis with antibodies to the indicated proteins. (C) Lysates of 786O cells stably expressing the indicated 3xFlag-tagged VHL derivatives were subjected to immunoblot analysis with antibodies to Flag, to HIF-2α, or to HSP90 (loading control). (D) Total RNA isolated from 786O cells stably expressing the indicated 3xFlag-tagged VHL derivatives was subjected to Northern blot analysis with a VEGF-specific probe. Ethidium bromide staining of 28S rRNA is also shown (loading control).
To examine whether the chimeric proteins VHL-M1 and VHL-M2 function as substrate-recognition subunits of E3s in vivo, we compared the abundance of HIF-2α, which is ubiquitylated by the VHL-Elongin B/C-Cul2-Rbx1 complex in 786O cells stably transfected with expression vectors for wild-type VHL (positive control) or for the VHL derivatives M1, M2, or M3 (a deletion mutant that lacks the VHL box, included as a negative control). As the previous report (Maxwell et al. 1999), forced expression of wild-type VHL in 786O cells resulted in a marked reduction in the abundance of HIF-2α, which is stable in mock-transfected 786O cells because of the lack of endogenous functional VHL protein (Fig. 4C). Expression of VHL-M1 or VHL-M2 induced a similar down-regulation of HIF-2α, whereas the amount of this protein was not affected by expression of VHL-M3. Consistent with these observations, Northern blot analysis of vascular endothelial growth factor (VEGF) mRNA, whose abundance is controlled by HIF-α, revealed that expression of wild-type VHL, VHL-M1, or VHL-M2, but not that of VHL-M3, reduced the amount of VEGF mRNA in 786O cells (Fig. 4D). Together, these observations suggest that the chimeric proteins composed of VHL and a Cul5 box form multiprotein complexes by assembly with Elongin B/C and the Cul5-Rbx2 module, and thereby mediate HIF-α ubiquitylation.
Identification of LRR-1 and FEM1B as VHL-box proteins that interact with Cul2-Rbx1
The observation that SOCS-box proteins associate with the Cul5-Rbx2 module, but not with Cul2-Rbx1, suggested the possibility that BC-box proteins other than VHL might also interact with the Cul2-Rbx1 module. To test this possibility, we subjected HEK293T cells to transient transfection with expression vectors for Cul2 tagged at its C terminus with the 3xFlag epitope and for Rbx1. Cul2-associated cellular proteins were purified by coimmunoprecipitation with anti-Flag, fractionated by SDS-PAGE, and stained with Coomassie blue (Fig. 5A). Mass spectrometric analysis of the coprecipitating proteins revealed that, in addition to VHL, the BC-box proteins LRR-1 and FEM1B (Fig. 1A) were associated with Cul2. We then transfected HEK293T cells with vectors for 3xFlag-tagged LRR-1 or FEM1B, and tested the abilities of the recombinant proteins to interact with the Cul2-Rbx1 module by coimmunoprecipitation experiments. LRR-1 and FEM1B were found to associate with the Cul2-Rbx1 module and with Elongin B/C, but not with Cul5-Rbx2, by both mass spectrometric (Fig. 5B) and immunoblot (Fig. 5C) analyses of the precipitates. Sequence alignment of VHL, LRR-1, and FEM1B revealed similarity in the region (designated the Cul2 box) downstream of the BC box (Fig. 1B). These results suggested that the Cul2 box might be responsible for binding of these proteins to the Cul2-Rbx1 module.
Figure 5.
Identification of LRR-1 and FEM1B as VHL-box proteins that interact specifically with endogenous Cul2-Rbx1. (A-C) Lysates of HEK293T cells transiently (A) or stably (B,C) expressing the indicated recombinant proteins were subjected to immunoprecipitation with anti-Flag, and the resulting precipitates were analyzed by SDS-PAGE and mass spectrometry (A,B) and immunoblotting (C) as described in Figure 2. (D) Domain organization of human wild-type VHL (VHL-WT) and the M4 mutant thereof, as well as of human wild-type LRR-1 (LRR-1-WT), and the M1 mutant thereof. BC-box (filled) and Cul2-box (hatched) are shown. (E,F) Lysates of HEK293T cells stably expressing the indicated recombinant proteins were subjected to immunoprecipitation with anti-Flag, and the resulting precipitates were analyzed by SDS-PAGE and mass spectrometry (E) and immunoblotting (F) as described in Figure 2. (G) Domain organization of human wild-type SSB4 (SSB4-WT) and the M1 mutant thereof. BC-box (filled) and Cul5-box (dotted) are shown. (H) Lysates of HEK293T cells stably expressing the indicated recombinant proteins were subjected to immunoprecipitation with anti-Flag, and the resulting precipitates were subjected to immunoblot analysis with antibodies to the indicated proteins.
The Cul2 box is required for the binding of VHL-box proteins to Cul2-Rbx1
To determine whether the Cul2 box is indeed required for the binding of VHL-box proteins to the Cul2-Rbx1 module, we constructed a VHL mutant (M4), in which residues 178-181 (LDIV) were each replaced with alanine, and examined its ability to bind to the Cul2-Rbx1 module (Fig. 5D). Although VHL-M4 associated with Elongin B/C, it did not interact with Cul2-Rbx1 (Fig. 5E,F). Similarly, a mutant (M1) of LRR-1, in which the residues of the Cul2 box (HIIP) corresponding to those mutated in VHL-M4 were also changed to alanine, did not bind to the Cul2-Rbx1 module (Fig. 5D-F), indicating that the Cul2 box is necessary for the interaction of VHL-box proteins with Cul2-Rbx1.
SSB4 chimeric protein containing a Cul2 box assembles with Cul2-Rbx1
To examine further whether the Cul2 box indeed determines specificity in binding to the Cul2-Rbx1 module, we constructed a mutant of SSB4 (M1) in which the Cul5 box (residues 246-273) was replaced with the Cul2 box of LRR-1 (residues 332-357) (Fig. 5G). Whereas wild-type SSB4 associated with Elongin B, Elongin C, Cul5, and Rbx2, SSB4-M1 interacted with Elongin B/C and Cul2-Rbx1 (Fig. 5H). We therefore conclude that the Cul2 box is an important determinant of the specificity of binding to the Cul2-Rbx1 module.
Reduced expression of the Cul2-Rbx1 module inhibits the degradation of HIF-2α
To investigate whether the VHL-mediated degradation of HIF-2α is dependent on the Cul2-Rbx1 pathway, we applied RNA interference (RNAi) to determine the effects of depletion of endogenous Cul2, Cul5, Rbx1, and Rbx2 in HeLa cells. Two independent sequences for each molecule were used as targets of RNA interference. Cells infected with a retroviral vector encoding either Cul2 (Cul2-1 or Cul2-2) or Cul5 (Cul5-1 or Cul5-2) shRNAs specific for Cul2 or Cul5 mRNAs exhibited a significant decrease in the abundance of Cul2 or Cul5, respectively, compared with that apparent in cells infected with a control vector for EGFP shRNA (Fig. 6A). Expression of EGFP, Cul5-1, or Cul5-2 shRNA did not affect HIF-2α expression, whereas that of Cul2-1 or Cul2-2 shRNA significantly increased the expression of HIF-2α (Fig. 6B). Similar effect was observed in cells infected with a retroviral vector encoding Rbx1 (Rbx1-1 or Rbx1-2) shRNAs specific for Rbx1 mRNA, whereas Rbx2 knockdown did not affect the abundance of HIF-2α (Fig. 6C,D). These results indicate that the VHL-mediated degradation of HIF-2α is dependent on Cul2-Rbx1, but not on Cul5-Rbx2 module.
Discussion
Protein degradation by the ubiquitin-proteasome pathway plays a fundamental role in determining the abundance of important regulatory proteins. In this pathway, E3s are thought to determine substrate specificity, and many diverse E3 molecules have been identified from database searches based mostly on amino acid sequence similarity to the HECT (Hershko and Ciechanover 1998), RING-finger (Joazeiro and Weissman 2000), or U-box (Aravind and Koonin 2000; Hatakeyama et al. 2001; Cyr et al. 2002) domains. ECS complexes constitute a subfamily of RING finger-type E3s and are well-characterized CDLs. Like F-box proteins in the SCF complex, SOCS-box proteins have been thought to serve as receptor subunits of the ECS complex. VHL, one of the earliest identified receptor subunits of the ECS complex, actually is atypical in that it lacks the C-terminal region of the SOCS-box domain that is conserved among SOCS-box proteins (Fig. 1) We now provide biochemical evidence that VHL and SOCS-box proteins specifically associate with the Cul2-Rbx1 and Cul5-Rbx2 modules, respectively, and that these interactions are determined by the presence of domains designated Cul2 and Cul5 boxes.
Our finding that Cul2 interacts with VHL, but not with SOCS-box proteins, raises the issue of whether VHL is the only substrate-recognition subunit of Cul2-Rbx1-based E3s. In Caenorhabditis elegans, the Cul2 homolog ceCUL2 is required for the G1-S transition of the cell cycle and for mitotic chromosome condensation (Feng et al. 1999), whereas inactivation of the human VHL gene results in constitutive activation of the hypoxic response, leading to the development of tumors such as clear cell renal carcinoma and cerebellar hemangioblastoma (Kaelin 2002). These observations suggest the existence of additional VHL-like proteins in mammalian cells that bind to the Cul2-Rbx1 module through the Elongin B/C adapter and function as substrate-recognition subunits. Indeed, we have now identified LRR-1 and FEM1B as Cul2-Rbx1-interacting proteins that contain a VHL box as well as protein-protein interaction motifs (leucine-rich repeats and ankyrin repeats, respectively). Like F-box proteins and SOCS-box proteins, a large number of unidentified VHL-box proteins might thus exist in cells and be responsible for recruiting specific substrates to Cul2-Rbx1-based E3s and positioning them in close proximity to the Cul2-Rbx1 module and the associated E2s.
Biochemical purification of VHL- and Cullin-interacting proteins led to the identification of the RING-finger protein Rbx1 in higher eukaryotes (Kamura et al. 1999; Ohta et al. 1999; Seol et al. 1999; Skowyra et al. 1999; Tan et al. 1999). With the subsequent help of database searches, we also identified the additional RING-finger protein Rbx2 (also known as Roc2) (Kamura et al. 1999; Ohta et al. 1999). Human Rbx1 and Rbx2 share an overall amino acid sequence identity of 51% and are equally effective in mediating the formation of polyubiquitin conjugates in association with multiple Cullin family members. However, it has remained unclear why such similar molecules exist and whether they are indeed completely functionally redundant or not. Drosophila contains three highly conserved _Rbx_- or _Roc_-like genes, one of which (Roc2) encodes a protein with 64% sequence identity to human Rbx2 and the other two of which (Roc1a, Roc1b) encode proteins that share 85% and 59% sequence identity with human Rbx1, respectively. Roc1a mutant cells accumulate Ci but not Arm, despite the requirement of the F-box protein Slimb for proteolysis of both Cubitus interruptus (Ci) and Armadillo (Arm) (Noureddine et al. 2002), suggesting that Slimb and Roc1a function in the same SCF complex to target Ci, but that a different Roc protein acts with Slimb to target Arm. This finding also indicates that the Drosophila Roc proteins are not functionally redundant. Recently, it was reported that the Drosophila Roc proteins preferentially bind to different members of the Cullin family; Roc1a binds to CUL-1, Roc1b binds to CUL-3, and Roc2 binds to CUL-5 (Donaldson et al. 2004). These observations suggest that each cullin selectively associates with a particular Rbx/Roc protein under the physiological condition. Our present data showing that Cul5 associated with SOCS-box proteins always forms a complex with Rbx2 (not with Rbx1) suggest that Rbx1 and Rbx2 in higher eukaryotes are also functionally distinct, at least in terms of their specific binding to Cullin family members.
Database searches have identified a large number of SOCS-box proteins (Masuhara et al. 1997; Hilton et al. 1998). Among the members of this family, however, only SOCS1 has previously been shown to promote the ubiquitylation of substrates, in this case JAK2, Vav, IRS1, and IRS2 (De Sepulveda et al. 2000; Kamizono et al. 2001; Rui et al. 2002). Although our present results now suggest that SOCS1 itself does not function as a component of an E3 because it contains the amino acid sequence IPLN in place of the conserved sequence LPXP in the Cul5 box, it is possible that the interaction of SOCS1 with these substrates triggers the recruitment of other E3s that actually mediate their ubiquitylation and degradation, or that SOCS1 weakly binds to the cullin-Rbx module. Replacement of the asparagine (N) residue in this sequence with proline (P) conferred on SOCS1 the ability to interact with the Cul5-Rbx2 module.
Many BC-box proteins are present in cells and regulate various cellular processes. These proteins can now be divided into three subfamilies on the basis of the proteins with which they interact. Those in the VHL-box and SOCS-box subfamilies form E3 complexes by interacting with Elongin B/C and either Cul2-Rbx1 or Cul5-Rbx2, respectively, and function as substrate-recognition subunits, whereas those in the third subfamily, such as Elongin A and Med8, bind to Elongin B/C, and thereby increase the stability of BC-box proteins (Kamura et al. 1998).
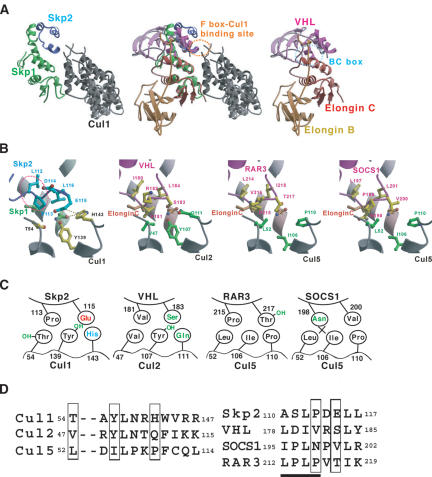
The crystal structure of the SCFSkp2 complex reveals that, in addition to Skp1, the F-box region of Skp2 directly binds to the N-terminal region of Cul1 (Fig. 7A; Zheng et al. 2002). In the loop region (amino acids 113-116) of Skp2, which is the binding interface with Cul1, Pro 113 participates in a hydrophobic interaction with Thr 54 of Cul1 and Glu 115 forms a hydrogen bond and a salt bridge with Tyr 139 and His 143 of Cul1, respectively (Fig. 7B). Given that the crystal structure of the VHL-Elongin B/C complex reveals a structural motif for Elongin C similar to that of Skp1, and that this motif appears to be responsible for Cul2 binding (Stebbins et al. 1999), the VHL-Elongin B/C-Cul2 complex can be superimposed onto the Skp2-Skp1-Cul1 complex in the view of this motif (Fig. 7A). From this structural comparison, it seems reasonable to assume that VHL may bind to Cul2 in a manner similar to that in which Skp2 interacts with Cul1 in the SCFSkp2 complex, suggesting that the loop region (amino acids 181-184) of VHL associates directly with Cul2. Furthermore, comparison of the amino acid sequences of Skp2, VHL, and SOCS-box proteins indicates that the putative Cullin-binding sites overlap with the LPXP motif of the SOCS box (Fig. 7C,D). Our present study demonstrates that the major determinant of Cullin-binding specificity of SOCS-box proteins is the LPXP motif of the SOCS box, especially the last proline of this motif. This terminal proline corresponds to Pro 113 of Skp2 (Val 181 of VHL), which directly contacts Thr 54 of Cul1. Schematic representation of the amino acids of the Cullin-binding interfaces (Fig. 7B,C) shows that Val 181 of VHL likely participates in a hydrophobic interaction with Val 47 of Cul2 and that Pro 215 of the LPXP motif of RAR3 interacts hydrophobically with Leu 52 of Cul5. However, Asn 198 of SOCS1, which corresponds to Pro 215 of RAR3, is not able to enter into a similar hydrophobic interaction with Cul5, consistent with our present findings that SOCS1 does not bind to Cul5, but that replacement of Asn 198 with Pro confers on SOCS1 the ability to bind to Cul5. The LPXP motif thus indeed appears to be an important determinant of Cullin-binding specificity.
Figure 7.
Structural comparison of putative Cullin-binding interfaces. (A) Comparison of the SCFSkp2 complex (left) and the VHL-Elongin B/C complex (right). Ribbon models of Cul1 (residues 17-355), Skp1, and Skp2 are shown in gray, green, and purple, respectively. Ribbon models of VHL (residues 62-209), Elongin B, and Elongin C are shown in magenta, orange, and red, respectively. Superposition of the VHL-Elongin B/C complex on the SCFSkp2 complex is also shown (middle) in the view of the similar structural motifs of Elongin C and Skp1. The F-box region of Skp2 directly binds to the N-terminal region of Cul1 (orange dotted circle). (B) The Cul1-binding interface of Skp2 and the putative corresponding site (Cul2 or VHL box) of VHL, RAR3 and SOCS1, each of which is highlighted by the orange dotted circle in A, are shown as ball-and-stick models. The red circle shown for the SCFSkp2 complex corresponds to the LPXP motif in the SOCS box. Color coding of proteins is the same as that in A. (C) Schematic representation of the amino acids at the Cullin-binding interfaces of Skp2, VHL, RAR3, and SOCS1. Hydrophilic, hydrophobic, and charged residues are shown in green, gray, and blue (positive) or red (negative), respectively. Threonine and tyrosine residues have both hydrophobic regions and hydrophilic hydroxyl groups, and are thus shown in gray and green (OH). Asp 198 of SOCS1 appears unable to participate in a hydrophobic interaction (cross). (D) Amino acid sequence comparisons among Cul1, Cul2, and Cul5 in the putative F-box, VHL-box, and SOCS-box-binding sites, respectively (left), as well as among Skp2, VHL, SOCS1, and RAR3 in the putative Cullin-binding sites. The LPXP motif of the SOCS box is underlined. Boxed residues are thought to participate in interactions. All residue numbers correspond to the human proteins.
We have identified LRR-1 and FEM1B as VHL-box proteins that interact with Cul2, and have provided direct biochemical evidence that the Cul2 box and the Cul5 box are required for the binding of VHL-box and SOCS-box proteins to the Cul2-Rbx1 or Cul5-Rbx2 modules, respectively. We thus propose to rename the Elongin B/C-Cul2-VHL-box protein and the Elongin B/C-Cul5-SOCS-box protein complexes as the ECV and ECS complexes, respectively. Together with the recent identification of BTB proteins as Cul3-associated proteins (Furukawa et al. 2003; Geyer et al. 2003; Pintard et al. 2003; Xu et al. 2003), our observations provide a foundation for future efforts to understand the biochemical events that are regulated by the Cullin-Rbx-based E3s.
Materials and methods
Antibodies
Mouse monoclonal antibodies (M2 and M5) to Flag were obtained from Sigma. A mouse monoclonal antibodies to Elongin C or to HSP90 were from Transduction Laboratories. Rabbit polyclonal antibodies to Cul5 (H-300) and a mouse monoclonal antibody to HIF-2α (190b), as well as goat polyclonal antibodies to Rbx2 (N-15) or to SOCS3 (M20) were from Santa Cruz Biotechnology. Rabbit polyclonal antibodies to Rbx1 were from Neomakers, those to Cul2 were from Zymed, those to SOCS3 (C005) were from Immuno-Biological Laboratories, and those to Elongin B were described previously (Garrett et al. 1995).
Construction of expression plasmids
The pcDNA3/Puro vector was constructed by replacing the neomycin resistance gene of pcDNA3 (Invitrogen) with the puromycin resistance gene. Complementary DNAs encoding wild-type or mutant versions of human SSB1, human SSB2, human SSB4, human RAR3, mouse WSB1, mouse MUF1, mouse SOCS1, human SOCS3, human VHL, human LRR-1, or human FEM1B, each tagged at their N termini with three copies of the Flag epitope (3xFlag), were subcloned into pcDNA3/Puro. A cDNA for human Cul2 tagged at its C terminus with 3xFlag was subcloned into pcDNA3. Mouse Rbx1 cDNA was subcloned into pCI-neo (Promega).
Transfection, immunoprecipitation, and immunoblot analysis
HEK293T cells or 786O cells were transfected with the indicated plasmids by the calcium phosphate method and selected in medium containing puromycin (10 μg/mL-1). Lysates (20 mg) of cells stably expressing the recombinant proteins were incubated for 1 h at 4°C with anti-Flag (M2) (100 μg) immobilized on NHS-activated Sepharose (Amersham Biosciences). The beads were then washed three times with a solution containing 40 mM Tris-HCl (pH 7.9), 150 mM NaCl, 1 mM dithiothreitol, and 0.5% Triton X-100, after which the immunoprecipitated proteins were eluted with the 3xFlag peptide (Sigma), separated by SDS-PAGE, transferred to a Hybond P membrane (Amersham Biosciences), and subjected to immunoblot analysis. Immune complexes were detected with Supersignal West Pico or West Dura chemiluminescence reagents (Pierce).
Mass spectrometric analysis
Immunoprecipitated proteins were separated by SDS-PAGE on an 8%-14% gradient gel and stained with Coomassie blue. The stained bands were excised from the gel and the proteins therein were subjected to in-gel reduction, S-carboxyamidomethylation, and digestion with trypsin. The resulting peptides were analyzed by LCQ ion-trap mass spectrometry (Aebersold and Mann 2003).
Northern blot analysis
Total RNA (7 μg) prepared with the ISOGEN reagent (Nippon Gene) was fractionated by electrophoresis on a 1% agarose gel, and the separated RNA molecules were transferred to a nylon membrane (Roche). A digoxigenin-labeled 750-bp DNA probe specific for human VEGF mRNA was generated by the polymerase chain reaction with a PCR DIG Probe Synthesis Kit (Roche). Hybridization was performed at 50°C for 5 h with the VEGF probe (0.1 mg/mL-1) in DIG Easy Hyb solution (Roche). The filter was then washed twice for 5 min at room temperature in 2× standard saline citrate (SSC) containing 0.1% SDS and then twice for 15 min at 60°C in 0.1× SSC containing 0.1% SDS. It was finally treated according to the protocol for a DIG Luminescent Detection Kit (Roche) with CDP-Star as a substrate and then exposed to RX-U film (Fujifilm). The agarose gel was also stained with ethidium bromide for comparison of the amount of 28S rRNA.
RNAi
The pMX-puro II vector was constructed by deletion of the U3 portion of the 3′ long terminal repeat of pMX-puro (kindly provided by T. Kitamura, University of Tokyo, Japan) (Morita et al. 2000). The mouse U6 gene promoter, followed by DNA corresponding to an shRNA sequence was subcloned into the NotI and XhoI sites of pMX-puro II, yielding pMX-puro II-U6/siRNA. The DNA for the shRNA encoded a 21-nucleotide hairpin sequence specific to the mRNA target, with a loop sequence (-TTCAAGAGA-) separating the two complementary domains, and contained a tract of five T nucleotides to terminate transcription. The hairpin sequences specific for human Cul2 (Cul2-1, Cul2-2), for human Cul5 (Cul5-1, Cul5-2), for Rbx1 (Rbx1-1, Rbx1-2), for Rbx2 (Rbx2-1, Rbx2-2), and for EGFP (Clontech) mRNAs corresponded to nucleotides 421-441 (Cul2-1), 1165-1175 (Cul2-2), 1637-1657 (Cul5-1), 2202-2222 (Cul5-2), 111-131 (Rbx1-1), 333-353 (Rbx1-2), 161-181 (Rbx2-1), 260-280 (Rbx2-2), and 126-146 (EGFP) of the respective coding regions. The resulting vectors were used to transfect Plat E cells and thereby to generate recombinant retroviruses. HeLa cells stably expressing mouse ecotropic retrovirus receptor were infected with the recombinant retroviruses. Four days after infection, cells were harvested and subjected immunoblotting.
Modeling of the Cullin-binding complexes
The crystal structures of SCFskp2 (Skp1/Skp2/Cul1) complex (Protein Data Bank accession no. 1LDK) (Zheng et al. 2002) and of VHL-Elongin B/C complex (Protein Data Bank accession no. 1vcb) (Stebbins et al. 1999) are used to provide a template for the Cullin/Cullin-binding protein docking geometry. The structure of a VHL-Elongin B/C complex was superimposed onto the SCFskp2 complex using as a basis the similar structural motif of the Elongin C and Skp1. In the template, the sequences of VHL/Cul2, RAR/Cul5, or SOCS1/Cul5 were substituted using the interactive graphics Program O (Jones et al. 1991). The appropriate Cullin-binding interfaces were modeled manually in Program O. Each superposition was performed with the program SHP (Stuart et al. 1979). Figure 7 was produced using Bobscript (Esnouf 1997) and was rendered with Raster3D (Merritt 1994).
Acknowledgments
We thank T. Kitamura for pMX-puro; J.M. Cunningham and K. Hanada for mCAT-1 plasmid; R. Yada, E. Fujimoto, and M. Oda for technical assistance; and M. Kimura and A. Ohta for help in preparation of the manuscript. This work was supported in part by grants from the Ministry of Education, Science, Sports, and Culture of Japan, and from the Human Frontier Science Program. We declare that we have no competing financial interests.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1252404.
References
- Aebersold R. and Mann, M. 2003. Mass spectrometry-based proteomics. Nature 422**:** 198-207. [DOI] [PubMed] [Google Scholar]
- Aravind L. and Koonin, E.V. 2000. The U box is a modified RING finger—A common domain in ubiquitination. Curr. Biol. 10**:** R132-R134. [DOI] [PubMed] [Google Scholar]
- Aso T., Lane, W.S., Conaway, J.W., and Conaway, R.C. 1995. Elongin (SIII): A multisubunit regulator of elongation by RNA polymerase II. Science 269**:** 1439-1443. [DOI] [PubMed] [Google Scholar]
- Aso T., Haque, D., Barstead, R.J., Conaway, R.C., and Conaway, J.W. 1996. The inducible elongin A elongation activation domain: Structure, function and interaction with the elongin BC complex. EMBO J. 15**:** 5557-5566. [PMC free article] [PubMed] [Google Scholar]
- Bai C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W., and Elledge, S.J. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86**:** 263-274. [DOI] [PubMed] [Google Scholar]
- Bradsher J.N., Jackson, K.W., Conaway, R.C., and Conaway, J.W. 1993. RNA polymerase II transcription factor SIII. I. Identification, purification, and properties. J. Biol. Chem. 268**:** 25587-25593. [PubMed] [Google Scholar]
- Cockman M.E., Masson, N., Mole, D.R., Jaakkola, P., Chang, G.W., Clifford, S.C., Maher, E.R., Pugh, C.W., Ratcliffe, P.J., and Maxwell, P.H. 2000. Hypoxia inducible factor-α binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 275**:** 25733-25741. [DOI] [PubMed] [Google Scholar]
- Cyr D.M., Hohfeld, J., and Patterson, C. 2002. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 27**:** 368-375. [DOI] [PubMed] [Google Scholar]
- De Sepulveda P., Ilangumaran, S., and Rottapel, R. 2000. Suppressor of cytokine signaling-1 inhibits VAV function through protein degradation. J. Biol. Chem. 275**:** 14005-14008. [DOI] [PubMed] [Google Scholar]
- Donaldson T.D., Noureddine, M.A., Reynolds, P.J., Bradford, W., and Duronio, R.J. 2004. Targeted disruption of Drosophila Roc1b reveals functional differences in the Roc subunit of Cullin-dependent E3 ubiquitin ligases. Mol. Biol. Cell 15**:** 4892-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D.R., Pause, A., Burgess, W.H., Aso, T., Chen, D.Y., Garrett, K.P., Conaway, R.C., Conaway, J.W., Linehan, W.M., and Klausner, R.D. 1995. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 269**:** 1402-1406. [DOI] [PubMed] [Google Scholar]
- Endo T.A., Masuhara, M., Yokouchi, M., Suzuki, R., Sakamoto, H., Mitsui, K., Matsumoto, A., Tanimura, S., Ohtsubo, M., Misawa, H., et al. 1997. A new protein containing an SH2 domain that inhibits JAK kinases. Nature 387**:** 921-924. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph Model 15**:** 132-134. [DOI] [PubMed] [Google Scholar]
- Feldman R.M., Correll, C.C., Kaplan, K.B., and Deshaies, R.J. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91**:** 221-230. [DOI] [PubMed] [Google Scholar]
- Feng H., Zhong, W., Punkosdy, G., Gu, S., Zhou, L., Seabolt, E.K., and Kipreos, E.T. 1999. CUL-2 is required for the G1- to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat. Cell Biol. 1**:** 486-492. [DOI] [PubMed] [Google Scholar]
- Furukawa M., He, Y.J., Borchers, C., and Xiong, Y. 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5**:** 1001-1007. [DOI] [PubMed] [Google Scholar]
- Garrett K.P., Aso, T., Bradsher, J.N., Foundling, S.I., Lane, W.S., Conaway, R.C., and Conaway, J.W. 1995. Positive regulation of general transcription factor SIII by a tailed ubiquitin homolog. Proc. Natl. Acad. Sci. 92**:** 7172-7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R., Wee, S., Anderson, S., Yates, J., and Wolf, D.A. 2003. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12**:** 783-790. [DOI] [PubMed] [Google Scholar]
- Guardavaccaro D. and Pagano, M. 2004. Oncogenic aberrations of cullin-dependent ubiquitin ligases. Oncogene 23**:** 2037-2049. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., Yada, M., Matsumoto, M., Ishida, N., and Nakayama, K.I. 2001. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276**:** 33111-33120. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover, A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67**:** 425-479. [DOI] [PubMed] [Google Scholar]
- Hilton D.J., Richardson, R.T., Alexander, W.S., Viney, E.M., Willson, T.A., Sprigg, N.S., Starr, R., Nicholson, S.E., Metcalf, D., and Nicola, N.A. 1998. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc. Natl. Acad. Sci. 95**:** 114-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30**:** 405-439. [DOI] [PubMed] [Google Scholar]
- Ivan M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J.M., Lane, W.S., and Kaelin Jr., W.G. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 292**:** 464-468. [DOI] [PubMed] [Google Scholar]
- Jaakkola P., Mole, D.R., Tian, Y.M., Wilson, M.I., Gielbert, J., Gaskell, S.J., Kriegsheim, A., Hebestreit, H.F., Mukherji, M., Schofield, C.J., et al. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292**:** 468-472. [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A. and Weissman, A.M. 2000. RING finger proteins: Mediators of ubiquitin ligase activity. Cell 102**:** 549-552. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47**:** 110-119. [DOI] [PubMed] [Google Scholar]
- Kaelin W.G. 2002. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2**:** 673-682. [DOI] [PubMed] [Google Scholar]
- Kamizono S., Hanada, T., Yasukawa, H., Minoguchi, S., Kato, R., Minoguchi, M., Hattori, K., Hatakeyama, S., Yada, M., Morita, S., et al. 2001. The SOCS box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J. Biol. Chem. 276**:** 12530-12538. [DOI] [PubMed] [Google Scholar]
- Kamura T., Sato, S., Haque, D., Liu, L., Kaelin Jr., W.G., Conaway, R.C., and Conaway, J.W. 1998. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes & Dev. 12**:** 3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T., Koepp, D.M., Conrad, M.N., Skowyra, D., Moreland, R.J., Iliopoulos, O., Lane, W.S., Kaelin Jr., W.G., Elledge, S.J., Conaway, R.C., et al. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284**:** 657-661. [DOI] [PubMed] [Google Scholar]
- Kamura T., Sato, S., Iwai, K., Czyzyk-Krzeska, M., Conaway, R.C., and Conaway, J.W. 2000. Activation of HIF1α ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. 97**:** 10430-10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T., Burian, D., Yan, Q., Schmidt, S.L., Lane, W.S., Querido, E., Branton, P.E., Shilatifard, A., Conaway, R.C., and Conaway, J.W. 2001. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276**:** 29748-29753. [DOI] [PubMed] [Google Scholar]
- Kibel A., Iliopoulos, O., DeCaprio, J.A., and Kaelin Jr., W.G. 1995. Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science 269**:** 1444-1446. [DOI] [PubMed] [Google Scholar]
- Masuhara M., Sakamoto, H., Matsumoto, A., Suzuki, R., Yasukawa, H., Mitsui, K., Wakioka, T., Tanimura, S., Sasaki, A., Misawa, H., et al. 1997. Cloning and characterization of novel CIS family genes. Biochem. Biophys. Res. Commun. 239**:** 439-446. [DOI] [PubMed] [Google Scholar]
- Maxwell P.H., Wiesener, M.S., Chang, G.W., Clifford, S.C., Vaux, E.C., Cockman, M.E., Wykoff, C.C., Pugh, C.W., Maher, E.R., and Ratcliffe, P.J. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399**:** 271-275. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. 1994. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 50**:** 869-873. [DOI] [PubMed] [Google Scholar]
- Morita S., Kojima, T., and Kitamura, T. 2000. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 7**:** 1063-1066. [DOI] [PubMed] [Google Scholar]
- Naka T., Narazaki, M., Hirata, M., Matsumoto, T., Minamoto, S., Aono, A., Nishimoto, N., Kajita, T., Taga, T., Yoshizaki, K., et al. 1997. Structure and function of a new STAT-induced STAT inhibitor. Nature 387**:** 924-929. [DOI] [PubMed] [Google Scholar]
- Noureddine M.A., Donaldson, T.D., Thacker, S.A., and Duronio, R.J. 2002. Drosophila Roc1a encodes a RING-H2 protein with a unique function in processing the Hh signal transducer Ci by the SCF E3 ubiquitin ligase. Dev. Cell 2**:** 757-770. [DOI] [PubMed] [Google Scholar]
- Ohh M., Park, C.W., Ivan, M., Hoffman, M.A., Kim, T.Y., Huang, L.E., Pavletich, N., Chau, V., and Kaelin, W.G. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2**:** 423-427. [DOI] [PubMed] [Google Scholar]
- Ohta T., Michel, J.J., Schottelius, A.J., and Xiong, Y. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3**:** 535-541. [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis, J.H., Willems, A., Johnson, J.L., Srayko, M., Kurz, T., Glaser, S., Mains, P.E., Tyers, M., Bowerman, B., et al. 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425**:** 311-316. [DOI] [PubMed] [Google Scholar]
- Reines D., Conaway, R.C., and Conaway, J.W. 1999. Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr. Opin. Cell. Biol. 11**:** 342-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L., Yuan, M., Frantz, D., Shoelson, S., and White, M.F. 2002. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J. Biol. Chem. 277**:** 42394-42398. [DOI] [PubMed] [Google Scholar]
- Seol J.H., Feldman, R.M., Zachariae, W., Shevchenko, A., Correll, C.C., Lyapina, S., Chi, Y., Galova, M., Claypool, J., Sandmeyer, S., et al. 1999. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes & Dev. 13**:** 1614-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91**:** 209-219. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Koepp, D.M., Kamura, T., Conrad, M.N., Conaway, R.C., Conaway, J.W., Elledge, S.J., and Harper, J.W. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284**:** 662-665. [DOI] [PubMed] [Google Scholar]
- Starr R., Willson, T.A., Viney, E.M., Murray, L.J., Rayner, J.R., Jenkins, B.J., Gonda, T.J., Alexander, W.S., Metcalf, D., Nicola, N.A., et al. 1997. A family of cytokine-inducible inhibitors of signalling. Nature 387**:** 917-921. [DOI] [PubMed] [Google Scholar]
- Stebbins C.E., Kaelin Jr., W.G., and Pavletich, N.P. 1999. Structure of the VHL-ElonginC-ElonginB complex: Implications for VHL tumor suppressor function. Science 284**:** 455-461. [DOI] [PubMed] [Google Scholar]
- Stuart D.I., Levine, M., Muirhead, H., and Stammers, D.K. 1979. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J. Mol. Biol. 134**:** 109-142. [DOI] [PubMed] [Google Scholar]
- Tan P., Fuchs, S.Y., Chen, A., Wu, K., Gomez, C., Ronai, Z., and Pan, Z.Q. 1999. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol. Cell 3**:** 527-533. [DOI] [PubMed] [Google Scholar]
- Tanimoto K., Makino, Y., Pereira, T., and Poellinger, L. 2000. Mechanism of regulation of the hypoxia-inducible factor-1 α by the von Hippel-Lindau tumor suppressor protein. EMBO J. 19**:** 4298-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wei, Y., Reboul, J., Vaglio, P., Shin, T.H., Vidal, M., Elledge, S.J., and Harper, J.W. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425**:** 316-321. [DOI] [PubMed] [Google Scholar]
- Zhang J.G., Farley, A., Nicholson, S.E., Willson, T.A., Zugaro, L.M., Simpson, R.J., Moritz, R.L., Cary, D., Richardson, R., Hausmann, G., et al. 1999. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl. Acad. Sci. 96**:** 2071-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Schulman, B.A., Song, L., Miller, J.J., Jeffrey, P.D., Wang, P., Chu, C., Koepp, D.M., Elledge, S.J., Pagano, M., et al. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416**:** 703-709. [DOI] [PubMed] [Google Scholar]