Thymic involution and rising disease incidence with age (original) (raw)
Significance
Understanding the risk factors of carcinogenesis is a major goal of biomedical research. Historically, the focus has been on the role of somatic mutations, and the reason for cancer typically occurring late in life is predominantly attributed to a gradual accumulation of such mutations. We challenge that view and propose that the decline of the immune system is the primary reason why cancer is an age-related disease. The immunological model featured here captures risk profiles for many cancer types and infectious diseases, suggesting that therapies reversing T cell exhaustion or restoring T cell production will be promising avenues of treatment.
Keywords: cancer, infectious disease, T cell, thymus, driver mutations
Abstract
For many cancer types, incidence rises rapidly with age as an apparent power law, supporting the idea that cancer is caused by a gradual accumulation of genetic mutations. Similarly, the incidence of many infectious diseases strongly increases with age. Here, combining data from immunology and epidemiology, we show that many of these dramatic age-related increases in incidence can be modeled based on immune system decline, rather than mutation accumulation. In humans, the thymus atrophies from infancy, resulting in an exponential decline in T cell production with a half-life of ∼16 years, which we use as the basis for a minimal mathematical model of disease incidence. Our model outperforms the power law model with the same number of fitting parameters in describing cancer incidence data across a wide spectrum of different cancers, and provides excellent fits to infectious disease data. This framework provides mechanistic insight into cancer emergence, suggesting that age-related decline in T cell output is a major risk factor.
T cells develop from hematopoietic stem cells as part of the lymphoid lineage and have the ability to detect foreign antigens and neoantigens arising from cancer cells. In the thymus, lymphoid progenitors commit to a specific T cell receptor and undergo selection events that screen against self-reactivity. Cells that pass these selection gates then leave the thymus, clonally expanding to form the patrolling naive T cell pool (1). The vast majority of vertebrates experience thymic involution (or atrophy) in which thymic epithelial tissue is replaced with adipose tissue, resulting in decreasing T cell export from the thymus. In humans, this is thought to begin as early as 1 y of age (2) (Fig. S1). The rate of thymic T cell production is estimated to decline exponentially over time with a half-life of ∼15.7 y (2–4), thereby following the function e−αt, with α = 0.044 y−1. Declining production of new naive T cells is thought to be a significant component of immunosenescence, the age-related decline in immune system function. With the recent successes of T cell–based immunotherapies (5), it is timely to assess how thymic involution may affect cancer and infectious disease incidence.
It is clear from epidemiological data that incidence of infectious disease and cancer increases dramatically with age, and, specifically, that many cancer incidence curves follow an apparent power law (6, 7). The simplest model to account for this assumes that cancer initiation is the result of a gradual accumulation of rare “driver” mutations in one single cell. Furthermore, the fitting of this power law model (PLM) can be used to estimate the number of such mutations (6, 7). Exponential curves (i.e., of the form e λt) have also been used to fit cancer incidence data (8), resulting in worse fits than the PLM overall. Nevertheless, it is worth noting that exponential rates close to α = 0.044 y−1 can be seen to emerge from the incidence data (Fig. S2), indicating the relevance of the thymic involution timescale. While the PLM fits well, it does not account for changes in the immune system with age. To better determine the processes underlying carcinogenesis, we asked whether an alternative model, based only on age-related changes in immune system function, might partly or entirely explain cancer incidence.
Results
Immunological Model.
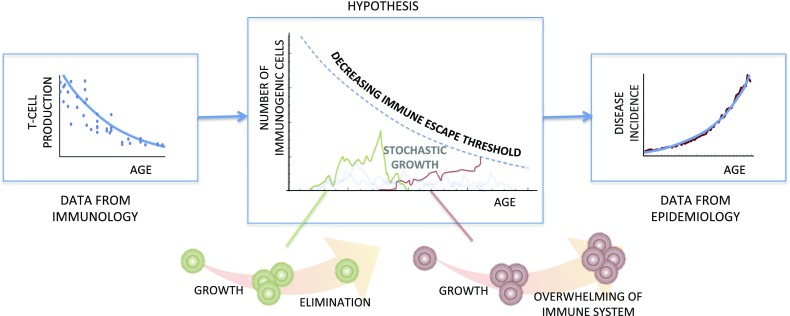
We developed a mathematical model of cancer incidence based on two assumptions: first, that potentially cancerous cells arise with equal probability at any age, and, second, that there exists an immune escape threshold (IET), proportional to T cell production, above which immunogenic cells can overwhelm the immune system and result in a clinically detectable disease (Fig. 1 and Fig. S3). For the sake of generality, as the model can also relate to age-related incidence of infectious diseases, the immunogenic cells could be mutated somatic cells or a population of infectious pathogens. We do not define the biological interaction between the T cell pool and the nascent tumor/infection; however, the concept of declining immune competence is consistent with several known mechanisms: for instance, both T cell repertoire diversity and the proliferative capacity of naive T cells decrease with age (9). Our model is thus derived as follows: once immunogenic cells arise, the population of such cells will change over time, leading to stochastic dynamics in population size, clonal diversity, and potentially other properties. The simplest way to capture these dynamics is through a birth–death process, and to a first approximation this can be modeled as a biased random walk (10). Fig. 1 provides a schematic view of the model dynamics in terms of population size. If the random walk exceeds the IET, the immune system will no longer be able to respond effectively and immune escape occurs.
Fig. 1.
Declining T cell production leads to increasing disease incidence. Our model assumes that immunogenic cells arise with the same probability at any age, and, after a period of being targeted, the population may overwhelm the immune system by crossing an immune escape threshold. This threshold is assumed to be proportional to T cell production, which decreases with age. This provides a prediction for the possible forms of disease incidence curves.
If the random walk for the immunogenic cells is unbiased (e.g., for the random walk describing population size, if cell division and cell death are equally likely), then the probability for an immunogenic cell population to reach a threshold K is given by 1/K (Methods). This gives a first-approximation prediction that the risk of immune escape, which we denote by R, rises exponentially with age at the same rate that T cell production declines. This defines a model for disease incidence with one fitting parameter, that being an overall prefactor (Table 1). If the random walk is biased (e.g., if the rates of cell division and cell death are not equal), a similar calculation produces a more general prediction for incidence with one additional parameter (Table 1). We will refer to these one- and two-parameter model predictions as, respectively, immune model I (IM-I) and immune model II (IM-II). The additional fitting parameter of IM-II can be interpreted as a “pivot age,” which marks a transition from very low to relatively much higher risk (Methods). We stress that α is not a fitting parameter, but the empirically derived rate from thymus involution, given by 0.044 y−1, which we use for all of our analysis.
Table 1.
Summary of the mathematical forms of the immune models and the PLM
| Model | Predicted risk profiles | Free parameters | Brief description |
|---|---|---|---|
| Immune model I | R=A eαt | A | Risk doubles every 16 y |
| Immune model II | R=A/(ee−α(t−τ)−1) | A, τ | Risk profile shifts around a pivot age of τ years |
| Power law (multistep) model | R=A tγ | A, γ | Waiting time for γ + 1 rare events (6, 7) |
Infectious Disease Incidence.
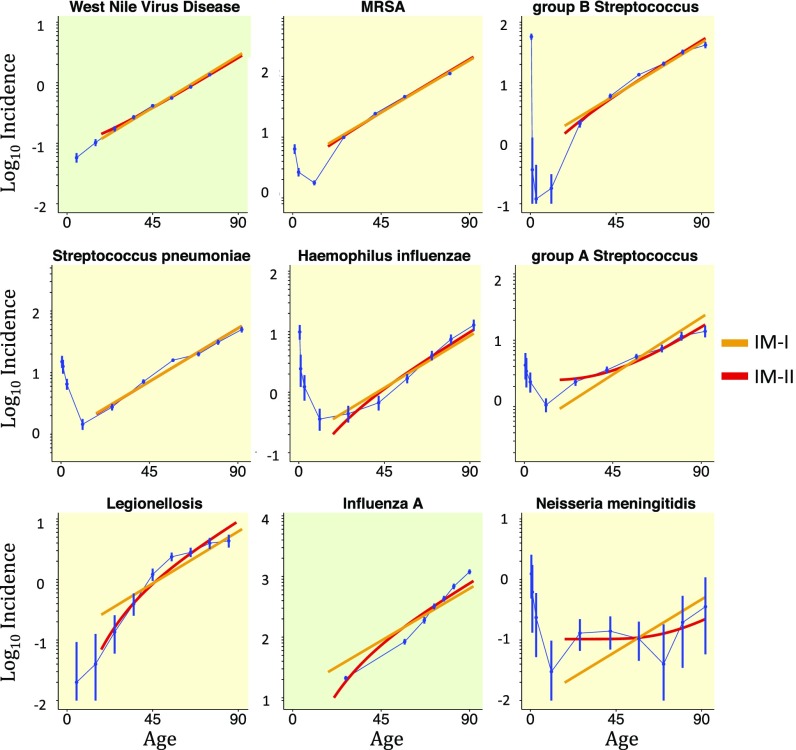
For most infectious diseases, the increase in risk with age is believed to be due to changes in the immune system and therefore provides a good first test for our model. The assumption that the immunogenic cells arise with equal probability at any age amounts to assuming constant exposure across age groups. We found that six of the seven bacterial infections monitored by the Active Bacterial Core (ABC) surveillance program (Data Sources) fit IM-II well (_R_2 > 0.9), with better fitting for those incidence curves underpinned by higher incidence and larger population sizes and hence associated with a smaller relative uncertainty [i.e., smaller confidence intervals (CIs)]. Turning to viral diseases, the incidence of West Nile virus (WNV) disease is particularly well fit by IM-II (and indeed IM-I). However, influenza A is not fit well, instead rising exponentially at a faster rate (Fig. 2). Prevalence of tuberculosis infection in Cambodia also fits the model well (Fig. S4). Indeed, even IM-I fits these infectious diseases very well, which confirms the importance of the thymic involution timescale. This provides confidence in applying our approach further.
Fig. 2.
Infectious disease incidence. Log-linear plots of incidence (per 100,000 person-years) by age group for all ABC bacterial infections, West Nile virus (WNV) disease, and Influenza A, ordered from best fit to worst. Bacterial and viral diseases are shaded yellow and green, respectively. The two-parameter IM-II is in red, while the one-parameter IM-I is in orange. Incidence often decreases initially from birth due to an underdeveloped immune system in infants; therefore, models are fitted only to data points for ages greater than 18 y. Error bars show 99% CIs for all diseases.
Cancer Incidence.
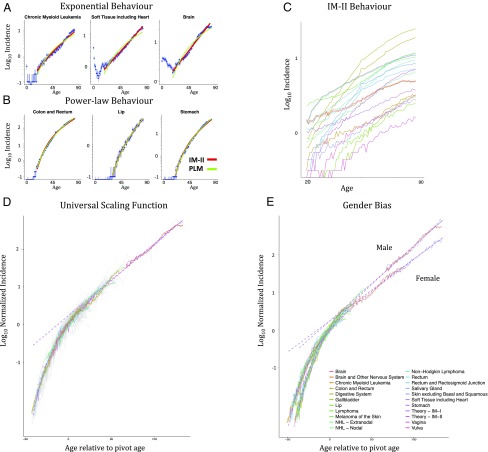
We next tested our model against cancer incidence curves, across 101 cancer types under the ICDO3 WHO2008 classification (11). Fitting IM-II to the incidence curves, the median _R_2 was found to be 0.956, with 57 cancer types fitting very well (_R_2 > 0.95). Since IM-II has the same number of fitting parameters as the widely used PLM of cancer incidence, a direct comparison is possible. The PLM performs slightly worse overall (_R_2 > 0.95 for 48 cancer types, median _R_2 = 0.947; _R_2 and associated fitting measures for each cancer type can be found in Dataset S1), with cancers whose incidence rises exponentially, such as chronic myeloid leukemia (CML) and brain cancer, fitting IM-I and IM-II better than the PLM. Many cancer types, including colon and gallbladder, fit both the PLM and IM-II very well (Fig. 3). There are no examples of PLM fitting well and notably better than IM-II. The ability of IM-II to capture the power law behavior seen in cancer incidence curves is an unexpected feature of the model and is discussed further in SI Theory [where we show that IM-II exhibits an apparent power law with power e/(e − 2) ∼ 3.78 in the age range of 33–82 y]. We note that, of the top 10 best-fit cancers, the 9 carcinomas have pivot ages tightly clustered from 56.3 to 60.5 y (Dataset S1), suggesting a clinical significance of the mid- to late fifties as an age of particular importance for screening and intervention. In contrast, the PLM by definition is “scale-free” and thus has no associated age range of particular importance from a clinical perspective.
Fig. 3.
Cancer incidence. Log-linear plots of incidence (per 100,000 person-years). Data taken from SEER (11). (A and B) Some cancer types rise exponentially fitting IM-I (A), while some cancer types rise like power laws, although can still be fit by IM-II (B). Fitting curves for IM-II and PLM are shown in red and green, respectively. (C) The top 20 best-fitting incidence curves as measured by Akaike Information Criterion (AIC) for IM-II. (D and E) Universal scaling functions for all cancers with defined pivot ages (84 out of 101 cancer types) plotted in gray with the top 20 incidence curves highlighted. Data shown for both genders (D) and gender-separated data (E), with dotted lines showing the model predictions for IM-I and IM-II. The gender-separated curves are fitted with higher independently determined values for α in males than females, reflecting the gender bias in T cell production (Methods). A purely exponential incidence curve would correspond to a pivot age of negative infinity, and therefore, for the purposes of plotting, we set a minimum pivot age of −50 y. Models are fitted only for ages greater than 18 y. Error bars show 95% CIs for all diseases.
From the form of the equation of IM-II we can see that, up to a shift in age and an overall multiplicative factor, all incidence curves should follow the same function (Methods and Fig. 3_D_). This “universal scaling function” shows the range of behaviors possible within the model. Indeed, the quality of data collapse of incidence data onto the universal scaling function for IM-II is excellent, giving strong support to our model and highlighting those cancer types that fit the model particularly well. One such cancer, CML, is characterized by a single translocation event resulting in the formation of the Philadelphia chromosome (12). This is a good candidate for the type of initiating event featured in our model. Assuming this translocation event can happen at any age, on neglecting the IET one might expect that incidence would be approximately constant. Instead, incidence doubles every 16 y, mirroring the exponential decay of T cell production, consistent with our model.
Examining which incidence curves fit poorly can give insight into the underlying diseases (Fig. S11, _R_2 < 0.9 for 28 cancer types with IM-II and 34 cancer types with PLM). For example, breast and thyroid cancer both rise rapidly and then plateau from middle age onward, possibly due to the significant hormonal influences for these cancers. Many cancer types have a plateau or even a dip in incidence around age 80. This cannot be explained by either IM-II or the PLM, since both give strictly increasing incidence with age. One can speculate that this decrease might be explained by declining tissue turnover. If this were the case, one would then expect cancer of the population of developing T cells itself (T cell lymphoblastic leukemia) to have an approximately constant risk profile with age, due to an exact cancellation of increasing risk from immunosenescence and decreasing risk from reduced cell production. This behavior is indeed observed when looking at adults above age 18 (Fig. S5). This finding supports the idea that both immune decline and decreasing tissue turnover contribute significantly to changes in cancer risk with age.
Our model has the potential to provide clinical insight into differences between cancer types. For example, those cancer types with a higher pivot age could be linked to tissues with a higher IET (Methods). This would decrease the probability of cancer initiation per se but would also imply that such cancers are larger or more advanced at the point of immune escape. From this, one would expect that pivot age should be inversely correlated with survivability, which indeed we observe (r = −0.6; value of P < 10−8; Fig. S7). To further test our model, we compared groups for which there are measurable differences in T cell production and disease incidence. While disease incidence is known to increase in immune-compromised groups, comparing males to females in the general population is more easily quantifiable. There is a gender bias in quantities of T cell receptor excision circle (TREC) DNA with age (4), which can be used to infer differences in naive T cell production between males and females. Interestingly, cancer is more common overall in males than females by a factor of 1.33 (13). We calculate that the TREC measurements from males and females have a similar gender bias, with females having 1.46 ± 0.31 (mean ± SD) more TREC DNA overall (4). As well as overall TREC counts, there is a difference in the rate of decline, with male TRECs falling faster (4). Consistently, the incidence data shows that 70 out of the 87 cancers with gender separation (i.e., observed in both genders) rise more steeply in males (Methods). To illustrate this bias, we constructed universal scaling functions for each gender (Fig. 3_E_ and Fig. S9_D_), each showing good data collapse. Interestingly, a similar gender bias is found in the average mutation burden in cancer biopsies, showing a steeper increase with age in males (14). WNV, the only infectious disease in our dataset with gender separation, also shows steeper increase in risk for males (value of P < 0.01).
For the majority of cancers, both the PLM and IM-II fit very well. To investigate further, we constructed a combined power law immunological model (PLIM), which includes rising risk with age from accumulating mutations and from immune system decline. This model has three fitting parameters, and is therefore relatively weaker as a predictive model, but does contain as submodels the PLM and IM-II (and hence IM-I). The model predicts risk profiles of the following form:
where A, B, and γ are fitting parameters (Methods). The parameter γ is once again interpreted as corresponding to γ + 1 driver mutations, while the parameter B indicates how much immune system decline contributes to rising risk with age. We found that this model provides good fits along a line in parameter space linking IM-II to the PLM (Fig. S8). For the cancers showing exponential behavior (brain, CML, and soft tissue including heart), the best fit is found very close to the IM-II (and indeed IM-I) region of parameter space, whereas for the cancers showing power law behavior, some cancers have their best fit close to the PLM region of parameter space. For colon and rectum cancer, the best fit occurs in-between the two regions, with a value of γ = 1.2 corresponding to 2.2 driver mutations. This estimate matches the value of 2.3 driver mutations for colorectal cancer found in ref. 15. Since the incidence curve fits a power law very accurately with exponent γ = 4.6, the estimate of 2.3 driver mutations suggests that the time dependence factorizes as R = _R_accumulation × _R_immune, where _R_accumulation ∝ _t_1.3 and _R_immune ∝ _t_3.3. It is noteworthy that the contribution of 3.3 to the net power law exponent of 4.6 is close to the value 3.78, which follows from the apparent power law behavior of IM-II, as discussed earlier. This factorization would imply that immune decline contributes more than accumulation of mutations to the increase in risk with age for colorectal cancer.
Discussion
We have shown that there is a strong link between T cell production and incidence of both infectious diseases and cancer. Some disease incidence curves rise exponentially, inversely proportional to T cell production (Fig. 3 and Fig. S8), while some rise in a manner well-captured by our two-parameter model, IM-II. This simple model, comprising (i) a threshold proportional to T cell production and (ii) a biased random walk characterizing the population dynamics of the immunogenic cells, can explain, to a large extent, cancer and infectious disease incidence, including gender differences. Further research is needed on the precise form of the IET to understand how it interfaces with declining T cell production in different diseases and individuals. The immunological model provides a fresh perspective on carcinogenesis, strongly supporting the idea that cancer can be caused by a single event in one cell that subsequently manages to beat the odds and evade the immune system through rare stochastic fluctuations in population dynamics. This is in stark contrast to the PLM, where the increase in risk with age arises from the waiting time for multiple independent events. We also predict that, for those animals that do not experience thymic involution, for example, some species of shark (16), cancer risk would not increase dramatically with age, and would thus be a relatively rare cause of death.
Mutations do indeed accumulate with age (17, 18), and although the premise of the PLM is logically and mathematically sound, this model predicts that several rare independent driver mutations are necessary for carcinogenesis. The fitted curves from the PLM and IM-II often overlap and can explain equally well many incidence curves. Further research is therefore necessary to estimate the number of driver mutations via other lines of inquiry. A recent paper (15) attempted to address this question from a new direction, by comparing groups with different mutation rates such as smokers and nonsmokers. Their analysis suggests that lung and colon cancer are caused by approximately n ∼ 2.3 driver mutations, rather than n ∼ 6.3 as would be inferred from the PLM alone. Our combined PLIM also predicts approximately n ∼ 2.2 driver mutations for colon cancer, although good fits (_R_2 > 0.95) are also found in other areas of parameter space. Moreover, a correlation has been found between the risk of cancer in a given organ and the total number of stem cell divisions estimated for that organ (19). It has been noted that this correlation does not show a highly nonlinear relationship, which would be expected from the mutation accumulation hypothesis (20). Indeed, if we apply the PLM to this dataset (Dataset S2), we find that the number of driver mutations is just n ∼ 0.91 (SI Theory) consistent with the assumptions underpinning the immunological model.
While IM-II has only two emergent fitting parameters, the underlying random-walk model has three biological parameters, resulting in an underdetermined system (Methods). To get estimates for these biological parameters, such as the size of the IET, additional assumptions are required. Given that the estimated total number of stem cell divisions provides a good predictor of cancer risk (19), the rate of stem cell divisions can be assumed to be proportional to the rate of cancer initiation attempts in the immunological model (SI Theory). From this, we can obtain values for the model parameters of IM-II. We found that the size of the IET is typically ∼106, which would imply that a population growing beyond 106 cancer cells would overwhelm the immune system and result in immune escape (see Dataset S1 for values for each cancer). In mouse experiments, primary inoculations with >106 cancer cells rendered mice unable to control subsequent tumor inoculations (21), providing a degree of qualitative and quantitative support for our model assumptions. This effect is related to the phenomenon of “T cell exhaustion,” which was initially defined as the clonal deletion of antigen-specific T cells due to chronic stimulation (22), and is now understood to involve not only activation-induced deletion but also changes in T cell phenotype and functionality (5). Therapies targeting T cell exhaustion have already been widely successful in cancer and infectious disease therapy in the form of immune checkpoint blockades such as PD-1 and CTLA-4 inhibitors (5). Our model provides a theoretical framework for such treatments and predicts that treatment efficacy could be enhanced if new naive T cell production were also increased. Additionally, evidence for a causative link between thymic activity and cancer risk has been found in mouse models, as thymectomized mice develop significantly more tumors (23, 24) and thymus grafts on nude mice can induce cancer remission (25, 26).
Our view supports the idea that as little as one single genomic aberration could be at the root of tumorigenesis. This event could be the emergence of a potent driver mutation, for example, a growth-inducing chromosomal translocation. Interestingly, it has been pointed out that a relatively small number of oncogenes have been confirmed across multiple biological experiments and all of these genes control cellular growth (27). Moreover, karyotypic analysis indicates that chromosomal rearrangements are encountered in most cancers in a way that is generally unique to the specific cancer under consideration (28). This led some to suggest that such changes are causative to cancer (29). Our analysis indicates that a single event (e.g., the emergence of a key mutation) could be enough to generate a malignancy that is able to evolve into a clinically manifest cancer if it escapes immune control. The immunological model also identifies a potential smoking gun in cancer risk in the form of the exponential decline of T cell production with age. Despite the decrease in T cell production from the thymus, overall T cell counts in the blood remain approximately constant due to increased peripheral clonal expansion (1). We therefore make the prediction that T cell efficacy is not increased by clonal expansion.
Our hypothesis and results add to the understanding of infectious disease and cancer incidence, suggesting in the latter case that immunosenescence, rather than gradual accumulation of mutations, serves as the predominant reason for an increase in cancer incidence with age for many cancers. For future therapies, including preventative therapies, strengthening the functionality of the aging immune system (30) appears to be more feasible than limiting genetic mutations, which raises hope for effective new treatments.
Methods
Immunological Model.
Simple models can often be very powerful in explaining complex phenomena (31, 32). With this in mind, we formulated a minimal model for disease incidence that does not attempt to explain the data exhaustively, but rather aims to be as simple as possible for the purposes of investigating the primary factors and rate-limiting steps.
During an immune response, immunogenic cells will be eliminated, while also increasing in number through division, such that the number of immunogenic cells follows a (biased) random walk. This stochastic birth–death process has been studied previously (10). The probability for reaching a population threshold K is given by the following:
where b and d are the birth (division) rates and death rates, respectively. The threshold K is interpreted as the largest number of immunogenic cells that can be effectively controlled by the immune system, and is thus the IET. Multiplying by the rate of initiating events r, we arrive at the predicted risk profile:
We assume that the only factor depending on age is K. The decrease of the IET with age is supported by experiments in mice showing a decline in proliferative capacity of activated T cells with age (33, 34). Specifically, we assume that the IET is proportional to the rate of export of naive T cells from the thymus. This would be the case if, for example, each T cell progenitor can only produce a finite number of daughter T cells and respond effectively to a finite maximum number of immunogenic cells, analogous to the Hayflick limit of replicative senescence (35). This gives K=K0 e−α t, leading to a predicted risk profile of the form R=A/(eBe−α t−1), where A = r(d − b)/b, B = _K_0 log(d/b).
Immunogenic cells are likely to have a higher division rate than normal cells, but since they are eliminated by the immune system, they will also have a higher death rate. Under the approximation that the division rate is equal to the death rate, Eq. 3 reduces to R=A′eα t, where A′ = r/_K_0. This constitutes a first-approximation prediction for risk profiles, with just a single fitting parameter.
When the fitting parameter B is negative, the biological parameters b and d satisfy b > d. In these rare cases, growth is approximately exponential and essentially a deterministic process, rather than a rare stochastic event. This would imply that the size of the threshold plays a small role and that incidence would be close to constant, which is indeed the case. For the majority of cases, especially the cases that can be fit well, the fitting parameter B is positive. To obtain a more easily interpreted model for these cases, we can repackage the parameter B and rewrite the full risk profile as follows:
where τ = log(B)/α. The parameter τ can now be interpreted as a pivot age, marking a change in behavior of the risk profile. For ages less than τ, the risk profile can be approximated as a steep Gompertz function R∼Ae−e−α(t−τ), while for ages greater than τ, the risk profile can be approximated as a pure exponential R∼Aeα(t−τ). In more biological terms, the pivot age represents the age when a cancer type transitions from very rare to relatively less rare. The median pivot age across all cancer types is τ = 49.9 y of age. The immune system’s response to a given cancer type influences the death rate d and also the immune exhaustion threshold size _K_0. In this way, a more competent immune system would lead to an increase in the pivot age parameter τ.
Up to a shift in age and an overall multiplicative factor, all functions of the form (6) can be collapsed onto a single universal scaling function given by the following:
where x = α(t − τ) and the overall multiplicative factor is chosen such that S(0) = 1. For the universal scaling function separated by gender, we have used values of exponent α higher in males than females. Since the available data on gender-separated TREC decline found in ref. 4 are very noisy (α for male TRECs is given by 0.08, with 0.05–0.11 95% CI, while α for female TRECs is given by 0.04, with 0.01–0.07 95% CI), we have arrived at values for α in males and females based on disease data. The cancer type which fits IM-I best is “soft tissue including heart.” This cancer has risk rising exponentially with exponents _α_M = 0.046 for males and _α_F = 0.038 for females, which we use for the universal scaling function. Consistently, the only infectious disease with gender separation, WNV, rises exponentially with exponents _α_M = 0.05 for males and _α_F = 0.041 for females.
The universal scaling function in Fig. 3 depicts the top 20 best-fitting cancers as measured by the Akaike information criterion (AIC). Other choices of measure give similar results (Fig. S10).
The immunological model above can be combined with the PLM to produce a model with three fitting parameters. To do so, we alter the assumption that potentially cancerous cells are produced at a constant rate, r, and assume instead that they arise from the gradual accumulation of driver mutations. Using the framework of the PLM (6, 7), the rate of attempts then takes the form r=r0tγ, corresponding to the waiting time for γ + 1 rare independent events. This PLIM predicts risk profiles of the following form:
where A = _r_0(d − b)/b, B = _K_0 log(d/b).
Data Sources.
Data sources for incidence rates are chosen based on largest possible sample sizes.
All cancer incidence data are obtained from Surveillance, Epidemiology, and End Results Program (SEER) in the United States (11).
Bacterial infection incidence data are obtained from the ABC surveillance program run by the Centers for Disease Control and Prevention (CDC). This program studies seven key bacterial diseases in detail (https://www.cdc.gov/abcs/reports-findings/surv-reports.html).
Incidence data for viral diseases is obtained from studies with the largest possible sample sizes. WNV disease incidence data are obtained from a 9-y survey covering the United States from 1999 to 2008 (available at https://www.cdc.gov/mmwr/preview/mmwrhtml/ss5902a1.htm; accessed February 23, 2016). Influenza A incidence data are obtained from a 22-y survey covering the United States (36).
Tuberculosis prevalence in Cambodia is obtained from ref. 37.
Stem cell counts and division rate estimates are taken from ref. 19.
Statistical Methods.
For incidence of infectious diseases and cancers, CIs are calculated assuming a χ2 distribution. All fitting of incidence curves is performed on log-transformed values.
To calculate the overall ratio of male TRECs to female TRECs, we computed the ratio of the means and then used a bootstrapping approach to calculate the SD of that measurement.
To show that cancer risk rises more steeply for males compared with females, we fit pure exponentials to the incidence curves and recorded the exponents as Female alpha and Male alpha in Dataset S1. To calculate the value of P for the statement that risk rises more steeply for WNV in males compared with females, we used the ANCOVA method.
All of the code for our analysis is available online at https://github.com/Albluca/ImmuneModelSEER.
Supplementary Material
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Acknowledgments
We thank Toni Aebischer, Md. Al Mamun, Doreen Cantrell, Mel Greaves, Sarah Howie, Philipp Kruger, Dianbo Liu, Luke McNally, Jacques Miller, Rob Newton, and Rose Zamoyska for useful discussions and comments on the manuscript. This work was supported by Scottish Universities Life Sciences Alliance and NIH through Physical Sciences in Oncology Centres Grant U54 CA143682 (to S.P., T.J.N., and L.A.), the Medical Research Council (C.C.B.), the European Union Seventh Framework Programme (FP7/2007–2013) collaborative project ThymiStem under Grant Agreement 602587 (to C.C.B.), and the Instituts Thématiques Multi-Organismes Cancer within the framework of the Plan Cancer 2014–2019 and convention Biologie des Systèmes BIO2014 (COMET project, to L.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.van den Dool C, de Boer RJ. The effects of age, thymectomy, and HIV Infection on alpha and beta TCR excision circles in naive T cells. J Immunol. 2006;177:4391–4401. doi: 10.4049/jimmunol.177.7.4391. [DOI] [PubMed] [Google Scholar]
- 2.Murray JM, et al. Naive T cells are maintained by thymic output in early ages but by proliferation without phenotypic change after age twenty. Immunol Cell Biol. 2003;81:487–495. doi: 10.1046/j.1440-1711.2003.01191.x. [DOI] [PubMed] [Google Scholar]
- 3.Douek DC, et al. Changes in thymic function with age and during the treatment of HIV infection. Nature. 1998;396:690–695. doi: 10.1038/25374. [DOI] [PubMed] [Google Scholar]
- 4.Sottini A, et al. Simultaneous quantification of T-cell receptor excision circles (TRECs) and K-deleting recombination excision circles (KRECs) by real-time PCR. J Vis Exp. 2014;2014:52184. doi: 10.3791/52184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang Y, Li Y, Zhu B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis. 2015;6:e1792. doi: 10.1038/cddis.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordling CO. A new theory on cancer-inducing mechanism. Br J Cancer. 1953;7:68–72. doi: 10.1038/bjc.1953.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br J Cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray F, Møller B. Predicting the future burden of cancer. Nat Rev Cancer. 2006;6:63–74. doi: 10.1038/nrc1781. [DOI] [PubMed] [Google Scholar]
- 9.Weng NP. Aging of the immune system: How much can the adaptive immune system adapt? Immunity. 2006;24:495–499. doi: 10.1016/j.immuni.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cisneros LH, Newman TJ. Quantifying metastatic inefficiency: Rare genotypes versus rare dynamics. Phys Biol. 2014;11:046003. doi: 10.1088/1478-3975/11/4/046003. [DOI] [PubMed] [Google Scholar]
- Surveillance, Epidemiology, and End Results (SEER) Program (2017) SEER*Stat Database: Incidence—SEER 18 Regs Research Data (2000–2016) (National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch). Available at https://seer.cancer.gov/. Accessed November 2, 2015.
- 12.Druker BJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 13.Dorak MT, Karpuzoglu E. Gender differences in cancer susceptibility: An inadequately addressed issue. Front Genet. 2012;3:268. doi: 10.3389/fgene.2012.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Podolskiy DI, Lobanov AV, Kryukov GV, Gladyshev VN. Analysis of cancer genomes reveals basic features of human aging and its role in cancer development. Nat Commun. 2016;7:12157. doi: 10.1038/ncomms12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomasetti C, Marchionni L, Nowak MA, Parmigiani G, Vogelstein B. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc Natl Acad Sci USA. 2015;112:118–123. doi: 10.1073/pnas.1421839112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shanley DP, Aw D, Manley NR, Palmer DB. An evolutionary perspective on the mechanisms of immunosenescence. Trends Immunol. 2009;30:374–381. doi: 10.1016/j.it.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Blokzijl F, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milholland B, Auton A, Suh Y, Vijg J. Age-related somatic mutations in the cancer genome. Oncotarget. 2015;6:24627–24635. doi: 10.18632/oncotarget.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomasetti C, Vogelstein B. 2015. Musings on the theory that variation in cancer risk among tissues can be explained by the number of divisions of normal stem cells. arXiv:1501.05035.
- 21.McBride WH, Howie SE. Induction of tolerance to a murine fibrosarcoma in two zones of dosage—the involvement of suppressor cells. Br J Cancer. 1986;53:707–711. doi: 10.1038/bjc.1986.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moskophidis D, Lechner F, Pircher H, Zinkernagel RM. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. 1993;362:758–761. doi: 10.1038/362758a0. [DOI] [PubMed] [Google Scholar]
- 23.Miller JF, Grant GA, Roe FJ. Effect of thymectomy on the induction of skin tumours by 3,4-benzopyrene. Nature. 1963;199:920–922. doi: 10.1038/199920a0. [DOI] [PubMed] [Google Scholar]
- 24.Anderson RE, Howarth JL, Troup GM. Effects of whole-body irradiation on neonatally thymectomized mice. Incidence of benign and malignant tumors. Am J Pathol. 1978;91:217–227. [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt M, Good RA. Transplantation of human cancers to nude mice and effects of thymus grafts. J Natl Cancer Inst. 1975;55:81–87. doi: 10.1093/jnci/55.1.81. [DOI] [PubMed] [Google Scholar]
- 26.Jacobsen GK, Povlsen CO, Rygaard J. Effects of thymus grafts in nude mice transplated with human malignant tumors. Exp Cell Biol. 1979;47:409–429. doi: 10.1159/000162960. [DOI] [PubMed] [Google Scholar]
- 27.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholson JM, Cimini D. Cancer karyotypes: Survival of the fittest. Front Oncol. 2013;3:148. doi: 10.3389/fonc.2013.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowak MA, et al. The role of chromosomal instability in tumor initiation. Proc Natl Acad Sci USA. 2002;99:16226–16231. doi: 10.1073/pnas.202617399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bredenkamp N, et al. An organized and functional thymus generated from FOXN1-reprogrammed fibroblasts. Nat Cell Biol. 2014;16:902–908. doi: 10.1038/ncb3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albergante L, Liu D, Palmer S, Newman TJ. 2016. Insights into biological complexity from simple foundations. Biophysics of Infection, Advances in Experimental Medicine and Biology, ed Leake MC (Springer, Berlin), pp 295–305. [DOI] [PubMed]
- 32.Al Mamun M, et al. Inevitability and containment of replication errors for eukaryotic genome lengths spanning megabase to gigabase. Proc Natl Acad Sci USA. 2016;113:E5765–E5774. doi: 10.1073/pnas.1603241113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joncourt F, Bettens F, Kristensen F, de Weck AL. Age-related changes of mitogen responsiveness in different lymphoid organs from outbred NMRI mice. Immunobiology. 1981;158:439–449. doi: 10.1016/S0171-2985(81)80014-9. [DOI] [PubMed] [Google Scholar]
- 34.Hobbs MV, et al. Cell proliferation and cytokine production by CD4+ cells from old mice. J Cell Biochem. 1991;46:312–320. doi: 10.1002/jcb.240460406. [DOI] [PubMed] [Google Scholar]
- 35.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 36.Thompson WW, Comanor L, Shay DK. Epidemiology of seasonal influenza: Use of surveillance data and statistical models to estimate the burden of disease. J Infect Dis. 2006;194(Suppl 2):S82–S91. doi: 10.1086/507558. [DOI] [PubMed] [Google Scholar]
- 37.Mao TE, et al. Cross-sectional studies of tuberculosis prevalence in Cambodia between 2002 and 2011. Bull World Health Organ. 2014;92:573–581. doi: 10.2471/BLT.13.131581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu S, Powers S, Zhu W, Hannun YA. Substantial contribution of extrinsic risk factors to cancer development. Nature. 2016;529:43–47. doi: 10.1038/nature16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spiess A-N, Neumeyer N. An evaluation of R2 as an inadequate measure for nonlinear models in pharmacological and biochemical research: A Monte Carlo approach. BMC Pharmacol. 2010;10:6. doi: 10.1186/1471-2210-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- 41.Willeit P, et al. Telomere length and risk of incident cancer and cancer mortality. JAMA. 2010;304:69–75. doi: 10.1001/jama.2010.897. [DOI] [PubMed] [Google Scholar]
- 42.Kuss I, et al. Recent thymic emigrants and subsets of naive and memory T cells in the circulation of patients with head and neck cancer. Clin Immunol. 2005;116:27–36. doi: 10.1016/j.clim.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 43.Yang H, et al. Obesity accelerates thymic aging. Blood. 2009;114:3803–3812. doi: 10.1182/blood-2009-03-213595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Youm Y-H, Horvath TL, Mangelsdorf DJ, Kliewer SA, Dixit VD. Prolongevity hormone FGF21 protects against immune senescence by delaying age-related thymic involution. Proc Natl Acad Sci USA. 2016;113:1026–1031. doi: 10.1073/pnas.1514511113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File
Supplementary File