Absence of Apparent Phenotype in Mice Lacking Cdc25C Protein Phosphatase (original) (raw)
Abstract
The Cdc25 family of protein phosphatases positively regulate the cell division cycle by activating cyclin-dependent protein kinases. In humans and rodents, three Cdc25 family members denoted Cdc25A, -B, and -C have been identified. The murine forms of Cdc25 exhibit distinct patterns of expression both during development and in adult mouse tissues. In order to determine unique contributions made by the Cdc25C protein phosphatase to embryonic and adult cell cycles, mice lacking Cdc25C were generated. We report that _Cdc25C_−/− mice are viable and do not display any obvious abnormalities. Among adult tissues in which Cdc25C is detected, its transcripts are most abundant in testis, followed by thymus, ovary, spleen, and intestine. Mice lacking Cdc25C were fertile, indicating that Cdc25C does not contribute an essential function during spermatogenesis or oogenesis in the mouse. T- and B-cell development was also found to be normal in _Cdc25C_−/− mice, and _Cdc25C_−/− mouse splenic T and B cells exhibited normal proliferative responses in vitro. Finally, the phosphorylation status of Cdc2, the timing of entry into mitosis, and the cellular response to DNA damage were unperturbed in mouse embryo fibroblasts lacking Cdc25C. These findings indicate that Cdc25A and/or Cdc25B may compensate for loss of Cdc25C in the mouse.
cdc25+ was initially identified as a temperature-sensitive allele that arrests fission yeast in G2 at the restrictive temperature (10). cdc25+ was cloned by functional complementation and shown to be a dose-dependent inducer of mitosis (42). In humans and rodents, there are three members of the Cdc25 family, designated Cdc25A, -B, and -C (13, 35, 36). Cdc25 family members are dual-specificity protein phosphatases that dephosphorylate and activate cyclin-dependent protein kinases (CDKs) (9, 16, 22, 33, 43, 47). Although all three human homologs can functionally complement a temperature-sensitive mutation of cdc25+ in fission yeast, each member is thought to contribute unique functions throughout embryonic development and in cycling somatic cells. In mice, Cdc25A, -B, and -C are expressed in overlapping but distinct patterns throughout development and show tissue-specific expression patterns in adult mice (27, 37, 51, 52).
Experiments performed using tissue culture cells have revealed several distinguishing characteristics of individual Cdc25 family members. Microinjection of Cdc25A antibodies arrests cells prior to S phase, and ectopic expression of Cdc25A shortens the G1 phase, thereby accelerating entry into S-phase (2, 21, 26, 44). These results suggest a role for Cdc25A at the G1-to-S-phase transition, although Cdc25A is active throughout all stages of the cell cycle (2, 21). The CDKs targeted by the Cdc25A protein phosphatase have not been definitively identified (2, 21, 23, 34, 44, 49). Microinjection of antibodies specific for either Cdc25B or Cdc25C arrests cells in G2, suggesting roles for these proteins at the G2-to-M-phase transition (32, 35). In certain cell types, Cdc25B has been shown to be an unstable protein that accumulates during the S- and G2-phases of the cell cycle, whereas in other cases the activity of Cdc25B is regulated so that it is most active during the S- and G2-phases of the cell cycle (12, 32, 38). Cdc25B has been shown to activate cyclin A-Cdk2 and cyclin B1-cdc2 complexes in vitro (11, 22, 43). Unlike Cdc25B, the intrinsic phosphatase activity of Cdc25C is low during the S and G2 phases of the cell cycle and is high in mitosis (8, 20, 25, 28, 30, 32, 50). Phosphorylation has been found to directly activate the enzymatic activity of Cdc25C, and Cdc2-cyclin B complexes appear to be the sole target of Cdc25C (9, 16, 20, 24, 25, 30, 33, 47). Two kinases have been shown to phosphorylate and activate Cdc25C in vitro. These include Cdc2-cyclin B1 and the Polo-like kinase Plx1 (20, 24, 29). It has been proposed that a positive feedback loop exists between Cdc2 and Cdc25C which rapidly induces both activation of Cdc2 and mitotic entry (20, 24, 46).
Cdc25A is primarily nuclear (21), whereas the localization of Cdc25B and Cdc25C varies as a function of the cell cycle. Both Cdc25B and Cdc25C shuttle in and out of the nucleus, and their accumulation in various subcellular compartments is influenced by nuclear localization sequences, nuclear export sequences, and protein-protein interactions (6, 7, 17, 18). All three human Cdc25 family members bind to 14-3-3 proteins, and the localization of both Cdc25C and Cdc25B is regulated by 14-3-3 interactions (5–7, 17, 18, 40).
Cdc25A and Cdc25B but not Cdc25C cooperate with an activated H-Ras mutant and with loss of retinoblastoma protein (RB) to transform primary rodent cells (14). Overexpression of Cdc25B has been observed in certain human cancer cell lines (36) as well as in primary human breast cancers (14). In addition, transgenic mice that overexpress Cdc25B in mammary epithelium more frequently develop carcinogen-induced mammary tumors (54). Both Cdc25A and Cdc25B are overexpressed in a certain percentage of head and neck cancers, non-small cell lung cancers, non-Hodgkin's lymphomas, and colorectal carcinomas (15, 19, 48, 53).
In summary, Cdc25 family members show distinct patterns of expression throughout development and in adult tissues of the mouse. In addition, individual Cdc25 family members can be distinguished by their intracellular localization, their abundance and/or activity throughout the cell cycle, and the cyclin or CDKs that they target for activation. Finally, Cdc25A and Cdc25B, but not Cdc25C, are proto-oncogenes. To further distinguish members of the Cdc25 family of protein phosphatases, we generated mice lacking Cdc25C by targeted gene disruption. We report that mice lacking Cdc25C are viable, develop normally, and do not display any obvious abnormalities.
MATERIALS AND METHODS
Construction of the Cdc25C targeting vector.
Cdc25C genomic clones were isolated from a 129/SvJ mouse embryonic stem (ES) cell genomic library (BAC-4921; Genome Systems, St. Louis, Mo.) by hybridization with murine Cdc25C (m_Cdc25C_)-specific cDNA probe (37). Restriction enzyme maps of the m_Cdc25C_ locus were determined using bacterial artificial chromosome (BAC) clones and genomic DNA from mouse tail clippings. One clone (BAC-35-F8) contained a 7-kb Eco_RV restriction fragment of genomic m_Cdc25C sequence, including exons 1 through 3 and ∼3.9 kb of sequence upstream of exon 1 and ∼1.1 kb of intronic sequence downstream of exon 3 (Fig. 1). The _Eco_RV genomic fragment was cloned into pBluescript-SK (Stratagene). The genomic organization of the mouse Cdc25C gene was disrupted by inserting the pTK-neo cassette (_Xba_I-_Hin_dIII fragment) derived from pSSC9 (4) into the Hpa_I site within exon 3 of m_Cdc25C. The cassette contains the neomycin phosphotransferase gene as a selectable marker flanked 5′ by the thymidine kinase promoter and 3′ by the thymidine kinase polyadenylation sequence. The targeting vector contained ∼5.9 kb and ∼1.1 kb of 5′ and 3′ homology region, respectively.
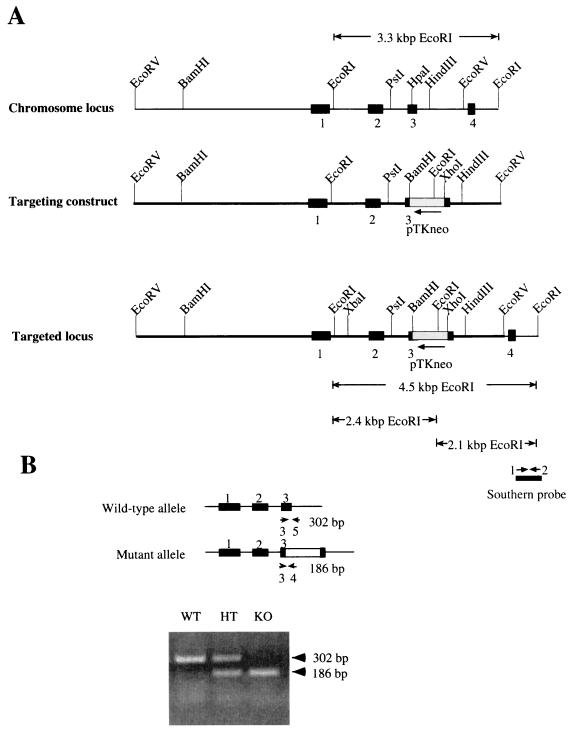
FIG. 1.
Targeted disruption of mouse Cdc25C gene. (A) Structure of the targeting vector. The genomic organization of the mouse Cdc25C gene was disrupted by inserting into exon 3 the neomycin phosphotransferase gene (neo) driven by the thymidine kinase promoter (pTKneo) as a selectable marker. Restriction sites in the introns and exons flanking the targeted exon are indicated. Exons 1 to 4 are represented by black boxes. The bar under the targeted locus indicates the probe used for Southern blot analysis. _Eco_RI digestion fragments probed by Southern blotting are also indicated. (B) PCR analysis of mouse tail DNA; PCR strategy and expected sizes of the amplified DNA fragments for wild-type (top) and Cdc25C mutant (bottom) alleles. Small arrows depict the locations of PCR primers used for genotyping. Wild-type (WT) mice produced a 302-bp PCR fragment, null mice (KO) generated a 186-bp fragment, and heterozygous mice (HT) gave rise to both the 302- and 186-bp PCR products.
Generation of mice harboring the Cdc25C mutation.
RW4 ES cells (Siteman Cancer Center at Washington University) were electroporated with _Sal_I-linearized targeting vector and selected with geneticin (G418; Gibco). For detailed procedures, see http://medicine.wustl.edu/∼escore. One hundred forty-four ES colonies resistant to G418 were analyzed for homologous recombination by Southern blotting using a PCR-generated probe corresponding to a 500-bp genomic DNA fragment located in intronic sequence between exons 4 and 5 (Fig. 1). The Southern probe was amplified by PCR using BAC DNA from clone BAC-35-F8 as a template with primer 1 (5′-GAGAGGGCCTCTTAACTG) and primer 2 (5′-CTTGCTGGGAAAGAAGCC). Wild-type (WT) mice produced a 3.3-kb _Eco_RI fragment, knockout (KO) mice generated a 2.1-kb _Eco_RI fragment, and heterozygous (HT) mice gave rise to both species. The _Xba_I-_Hin_dIII fragment of pSSC9 containing the neomycin phosphotransferase gene was used as a probe to ensure that random integration of the targeting vector had not occurred elsewhere in the genome. Four ES clones heterozygous for Cdc25C were injected into C57BL/6 blastocysts, which were subsequently implanted into the uteri of pseudopregnant C57BL/6 × C3HF1 foster mothers. Male chimeras from three clones selected by agouti color were mated to C57BL/6 females. Germ line transmission was obtained for two clones. F1 animals were tested for the targeted Cdc25C allele by Southern blotting and PCR analysis of tail DNA. PCR analysis was achieved using a three-primer PCR with primer 3 (5′-GGTTCCTTGGATTCATCTGGACC) from exon 3, primer 4 (5′-CCTCGTGCTTTACGGTATCGCCG) from the neomycin cassette, and primer 5 (5′-CCCTACCATGAGTGCAGGGCACC) from intronic sequence between exons 3 and 4 (Fig. 1B). Heterozygous F1 males and females were interbred to generate F2 littermates used for subsequent breeding and analysis.
Histology.
For histological studies, tissues were fixed in 10% neutral buffered formalin, rinsed in phosphate-buffered saline (PBS), and stored in 70% ethanol. Fixed tissues were embedded in paraffin by standard procedures. Blocks were sectioned (5 μm) and stained with hematoxylin and eosin.
Animal growth measurements.
Heterozygous crosses were used to generate mice, and postnatal growth curves were obtained by weighing pups at the indicated times on a Mettler AE 50 scale. Genotyping was performed by PCR analysis of tail cuttings. All recorded weight averages shown in the growth curves were compiled from at least 30 mice per genotype per time point.
Purification and stimulation of B and T cells for Northern analysis.
Splenic and thymic tissues were homogenized in PBS with 1% fetal calf serum (FCS) using frosted glass slides. Red blood cells were removed by density gradient centrifugation using Histopaque-1119 (Sigma Chemical Company). Cells were pelleted, washed, and resuspended in PBS with 1% FCS. CD4+ T cells were isolated using Dynabeads Mouse CD4(L3T4) and detached from the beads using DETACHaBEAD mouse CD4 according to the manufacturer's instructions (Dynal). The remaining cells were ∼50% B220+ B cells. A total of 2 × 106 cells were plated in each well of a six-well Costar tissue culture dish. T and B cells were stimulated for 72 h with concanavalin A (5 μg/ml) and lipopolysaccharide (1 μg/ml), respectively (both from Sigma Chemical Co.).
Northern blotting.
RNA was isolated from cells and tissues using the QuickPrep total RNA extraction kit (Amersham Pharmacia Biotech). Testes were frozen in liquid nitrogen, suspended in tissue extraction buffer (QuickPrep total RNA extraction kit from Amersham Pharmacia Biotech), and homogenized using a PowerGen 700 (Fisher Scientific). T- and B-cell populations were suspended in lithium chloride buffer prior to the extraction buffer. RNA was isolated according to the manufacturer's suggestions. RNA was resolved on a 1.2% agarose gel and then transferred to GeneScreen Plus (NEN). The probes used for screening mouse tissues and cells include a 440-bp _Hin_cII-Dra_III restriction fragment of the m_Cdc25C cDNA; a 427-bp Xmn-Pvu_I restriction fragment of the m_Cdc25A cDNA; a 466-bp _Bgl_II-Sac_II restriction fragment of the m_Cdc25B cDNA; a 600-bp _Hin_dIII-_Eco_RI restriction fragment of the glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH); and a ∼1.2-kb _Xba_I-_Hin_dIII restriction fragment of the neomycin phosphotransferase gene. Probes were labeled with [_α_-32P]dCTP (NEN) using the Megaprime DNA labeling system (Amersham). Blots were prehybridized with ExpressHyb solution (Clontech) containing sonicated salmon sperm DNA (100 μg/ml) for 2 to 3 h at 68°C with shaking. Labeled probe was added to 106 cpm/ml, and the blot was hybridized in ExpressHyb solution for 2 h at 68°C. The blot was then rinsed briefly with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate (SDS) at 65°C twice and then washed once with 2× SSC–0.5% SDS for 15 min with shaking at 65°C.
Generation of MEFs.
Mouse embryo fibroblasts (MEFs) were derived from 13.5-day-old embryos. Following removal of the head and organs, embryos were rinsed with PBS, minced, and digested with trypsin-EDTA (0.5% trypsin, 0.53 mM EDTA) for 10 min at 37°C, using 1 ml per embryo. Trypsin was inactivated by addition of Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL) containing 10% fetal bovine serum (FBS), 2 mM l-glutamine, 0.1 mM nonessential amino acids, and 140 μM 2-mercaptoethanol plus 100 U of penicillin G and 100 μg of streptomycin per ml. Cells from single embryos were plated into one 100-mm diameter tissue culture dish and incubated at 37°C in a 10% CO2 humidified chamber for 3 days. Cells were trypsinized, and 9 × 105 cells were replated into 60-mm tissue culture dishes every 3 days. Each trypsinization and replating represented one passage. Early-passage cells (P4, prior to crisis) were analyzed for cell cycle progression and for cellular responses to DNA damage. Later-passage, immortalized cells were used for the 35S-labeling experiments.
Antibodies.
mCdc25C was detected with a rabbit polyclonal peptide antibody (sp1573) raised against 14 amino acids within the C terminus of mCdc25C (CQLQGQIALLVKGAS) coupled to keyhole limpet hemocyanin or a rabbit polyclonal antibody raised against bacterially produced glutathione _S_-transferase (GST)-mCdc25C. GST fusion proteins were detected with a rabbit polyclonal antibody specific for GST (Z-5; Santa Cruz Biotechnology). Cdc2 protein was detected with Cdc2-specific rabbit polyclonal antibody (39). Bound primary antibodies were detected with horseradish peroxidase (HRP)-conjugated rat anti-rabbit immunoglobulin (Ig) antibody (Zymed), and proteins were visualized by chemiluminescence using the ECL reagent (Amersham).
Specificity of peptide antibody.
All procedures relating to Sf9 insect cell culture and propagation of recombinant baculoviruses were performed as described elsewhere (41). Baculoviruses encoding GST fusion proteins of murine Cdc25A, -B, and -C were generated using the Bac-to-Bac baculoviral expression system (Gibco-BRL) and vector pFASTBAC1 by standard procedures (41). Sf9 insect cells were infected with recombinant baculoviruses, and 45 h after infection, cells were washed with PBS and lysed in mammalian cell lysis buffer (MCLB: 50 mM Tris-HCl [pH 8.0], 2 mM dithiothreitol, 5 mM EDTA, 0.5% Nonidet P-40, 150 mM NaCl, 1 μM microcystin, 1 mM sodium orthovanadate, 2 mM phenylmethylsulfonyl fluoride, 0.15 units of aprotinin per ml, 20 μM leupeptin, and 20 μM pepstatin), and 100 μg of total cellular protein was resolved on an SDS–7% polyacrylamide gel, transferred to nitrocellulose membranes, and analyzed for Cdc25A, -B, and -C using a rabbit polyclonal antibody specific for GST or a peptide antibody specific for mCdc25C.
Binding of peptide antibody to immobilized protein A.
ImmunoPure immobilized protein A-agarose (Pierce) and sp1573 sera were incubated for 1 h at room temperature in 1 ml of MCLB at a ratio of 1 μl of antiserum to 2.5 μl of packed beads. The antibody-bead mixture was washed twice with 1 ml of MCLB, twice with 1 ml of LiCl buffer (0.5 M LiCl, 50 mM Tris[pH 8.0]), and twice with 1 ml of MCLB. For peptide competition experiments, 25 μl of packed antibody-coupled protein A-agarose was incubated in 100 μl of MCLB containing 2 μM peptide for 30 min at room temperature.
35S labeling and mCdc25C immunoprecipitations.
Tissue culture dishes (100 mm) were seeded with 2 × 106 MEFs 1 day prior to labeling. Cells were rinsed with 5 ml of labeling medium (DMEM lacking methionine and cysteine) and then incubated with labeling medium supplemented with 10% dialyzed FBS for 30 min. [35S]methionine-cysteine (NEN) was added to a final concentration of 0.5 mCi/ml, and cells were incubated overnight. Cells were washed with PBS and harvested by trypsinization. Cells were lysed in MCLB at 4°C for 15 min, and lysates were centrifuged at 10,000 × g for 10 min at 4°C. Equal amounts of trichloroacetic acid-precipitable counts from Cdc25C+/+ and _Cdc25C_−/− MEFs representing ∼3 mg of total cellular protein were precleared by incubation with 25 μl of packed ImmunoPure immobilized protein A per sample for 2.5 h at 4°C. Precleared lysates were then incubated with 25 μl of packed antibody-bead mixture (prepared as described above) for 3 h at 4°C. Immunoprecipitates were centrifuged and washed twice with LiCl buffer and four times with MCLB. Washed beads were resuspended in 0.2 ml of SDS lysis buffer (0.4% SDS, 100 mM NaCl, 50 mM Tris [pH 7.4], 2 mM EDTA), boiled for 5 min, and then cooled to room temperature, and 40 μl of buffer T (10 mM Tris [pH 7.5] 10 mM NaCl, 1 mM EDTA, 10% Triton X-100) was added. The mixture was clarified by centrifugation for 10 min. The clarified supernatant was transferred to a fresh tube, and 0.5 volume of MCLB was added. Cdc25C was reimmunoprecipitated with 25 μl of packed antibody-bead mixture at 4°C overnight. Immunoprecipitates were washed four times in MCLB and then resolved by SDS–7% polyacrylamide gel electrophoresis (PAGE). The gel was treated with 1 M sodium salicylate for 30 min, and labeled proteins were visualized by fluorography (3).
Cell cycle analysis.
MEFs at passage 4 (106) were seeded onto 100-mm tissue culture dishes 36 h prior to bromodeoxyuridine (BrdU) labeling. Cells were incubated in 5 ml of culture medium (DMEM with 10% FBS and 0.1 mM nonessential amino acids) containing 20 μM BrdU (Amersham) at 37°C for 1 h. The medium was removed and replaced with 3 ml of culture medium, and cells were either mock irradiated or exposed to 4 Gy from a 60Co source. An additional 7 ml of culture medium was added, and cells were incubated for the indicated times. Cells were harvested by trypsinization and collected by centrifugation. After removal of the supernatant by aspiration, cells were washed once in PBS and then suspended in 0.5 ml of PBS. Cells were fixed by the addition of 5 ml of 70% ethanol at 4°C in the dark. Pelleted cells were resuspended in 1 ml of 0.4-mg/ml pepsin (Sigma Chemical Co.) in 0.1 N HCl and incubated with rocking for 30 min. Nuclear pellets were suspended in 1 ml of 2N HCl–0.5% NP-40 and incubated with rocking for 1 h. After neutralization by incubation with 1 ml of 0.1 M sodium borate (pH 8.5) for 5 min, nuclear pellets were suspended in 100 μl of PBS-TB (PBS with 0.5% Tween 20 and 1% bovine serum albumin) and stained with 1.4 μg of fluorescein isothiocyanate (FITC)-conjugated anti-BrdU monoclonal antibody (Caltag Inc.) for 1 h in the dark. Nuclei were washed once with 1 ml of PBS-TB and then incubated with 1 ml of PBS-TB containing propidium iodide (PI, 30 μg/ml) for 30 min in the dark. Cells were analyzed for DNA content by fluorescence-activated cell sorting (FACS) using a FACS Calibur (Becton Dickinson Instruments). The data were analyzed using CellQuest analysis software (Becton Dickinson).
Synchronization of MEFs and analysis of Cdc2 phosphorylation.
MEFs (5 × 105) at passage 6 were seeded onto 100-mm tissue culture dishes. The following day, cells were incubated in 10 ml of culture medium (DMEM with 2 mM l-glutamine and 0.1 mM nonessential amino acids plus 100 U of penicillin G and 100 μg of streptomycin per ml) containing 0.1% FBS for 48 h. Cells were then incubated in culture medium containing 15% FBS and aphidicolin (1 μg/ml) (Calbiochem) for 20 h. Cells were released from the aphidicolin block by washing twice with 10 ml of PBS and then incubating in culture medium containing 15% FBS. Cells were harvested prior to release (time 0) or at 3, 6, 9, or 12 h after release and analyzed for DNA content or processed to monitor for Cdc2 phosphorylation status. For DNA content analysis, cells were suspended in 0.5 ml of PBS, fixed by the addition of 5 ml of 70% ethanol, and then stained with PI. Analyses were performed on a Becton Dickinson FACS Calibur equipped with CellQuest software. For monitoring Cdc2 phosphorylation, cells were lysed in MCLB for 10 min on ice. Cell lysates containing 2 mg of total cellular protein were incubated with 50 μl of packed p13_suc1_-agarose (Upstate Biotechnology). After incubation at 4°C for 3 h, precipitates were washed four times with 1 ml of MCLB. Proteins were resolved by SDS-PAGE on a 10% gel. Cdc2 phosphorylation status was monitored by immunblotting.
Flow cytometry of immune cells.
Splenic and thymic tissues were homogenized in PBS with 1% FCS using frosted glass slides. Red blood cells were removed by density gradient centrifugation using Histopaque-1119 (Sigma Chemical Company). Cells were pelleted, washed, resuspended in PBS with 1% FCS, and counted using a hemacytometer. A total of 106 cells were mixed with various monoclonal antibodies conjugated to either FITC or phycoerythrin (PE) from PharMingen. Antibodies included FITC-conjugated anti-CD4, PE-conjugated anti-CD8, PE-conjugated anti-CD45R/B220, and FITC-conjugated anti-CD3. All antibodies were used at 1 μg/106 cells after blocking nonspecific Fc binding with anti-CD16/CD32 cocktail (PharMingen). Analyses were performed on a Becton Dickinson FACS Calibur equipped with CellQuest software.
Proliferation assays.
Proliferation assays were performed with purified populations of CD4+ T cells and B220+ B cells. Splenic tissue was homogenized in K5 medium (RPMI 1640 containing 10% FBS, 15 mM HEPES, 10 μM nonessential amino acids, 1 mM sodium pyruvate, 2 mM l-glutamine, 50 nM 2-mercaptoethanol, 100 U of penicillin/ml, 100 μg of streptomycin/ml) using frosted glass slides. Red blood cells were removed by density gradient centrifugation using Histopaque-1119 (Sigma Chemical Company). Cells were pelleted, washed, and resuspended in K5 medium. CD4+ T cells were isolated by positive selection using Dynabeads Mouse CD4(L3T4) and detached from the beads using DETACHaBEAD mouse CD4 according to the manufacturer's instructions (Dynal). CD4+-depleted cells were next incubated with MACs beads (Miltenyi Biotech) at 10 μl of magnetic beads/107 cells to specifically bind CD43-positive cells. B cells that flowed through the column were used for proliferation assays. Purified B cells and CD4+ T cells (2 × 105) were plated in each well of a 96-well Costar tissue culture dish. Cells were incubated for the indicated amounts of time. CD4+ T cells were grown on plate-bound anti-CD3 monoclonal antibody (0.1 to 2 μg/ml; PharMingen). Goat anti-mouse IgM F(ab′)2 (Jackson Immunoresearch) was used as the B-cell mitogen at a final concentration of 20 μg/ml. All conditions were tested in triplicate. Cells were incubated with 2 μCi of [3H]thymidine per well (10 μCi/ml final) during the final 12 h of culture. Cells were harvested using a Skatron Instruments Micro96 harvester and counted using a Beckmann 6500 multipurpose scintillation counter.
RESULTS AND DISCUSSION
Targeted disruption of Cdc25C gene.
Genomic clones of the murine Cdc25C gene obtained from a 129/SvJ mouse ES cell library were used for construction of the pKO_Cdc25C-neo_ targeting construct (Fig. 1A). pKO_Cdc25C-neo_ was used to disrupt exon 3 in the endogenous mouse Cdc25C gene by homologous recombination. In addition, the construct was designed so that if splicing were to occur between exons 2 and 4 to remove the neo cassette, the resulting transcript would not be in frame. The targeted Cdc25C allele was introduced into the mouse genome by electroporation into the RW4 ES cell line, which was derived from mouse strain 129/SvJ. Of 144 G418-resistant clones screened for homologous recombination by Southern blot analysis, 5 (3%) contained correct targeting events. Four of the targeted clones were injected into C57BL/6 blastocysts, and a total of 18 chimeric males from four independent clones were obtained. Two of the four clones produced germ line-transmitting chimeric males. Southern blot and PCR analysis indicated that ∼50% of the agouti offspring produced by chimeric males were heterozygous for the targeted mutation of the Cdc25C locus. Mice from two independent targeted ES cell lines were separately bred to homozygosity for the disrupted Cdc25C gene. Results reported were observed in both lineages.
F1 heterozygous offspring were intercrossed, and F2 offspring were genotyped by Southern blotting (data not shown) or by PCR analysis. As shown in Fig. 1B, all three genotypes were detected. Cumulative genotyping of heterozygous crosses revealed that the numbers of wild-type to heterozygous to homozygous mutant mice were 197 (23%) to 444 (51%) to 229 (26%). Similar ratios were obtained when males and females were tallied separately (data not shown).
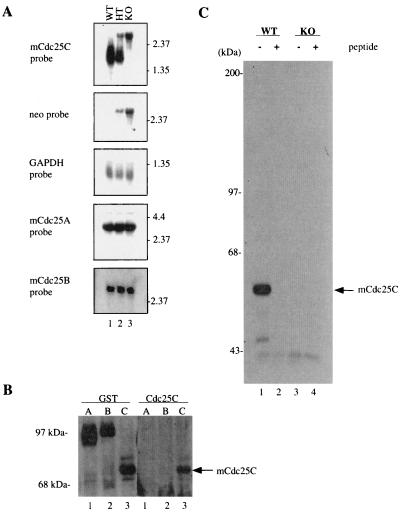
Mice were examined for the presence of Cdc25C at both the RNA and protein levels to verify that the targeted disruption of Cdc25C was successful in producing a null allele of the locus. Northern blot analysis was performed on total testicular RNA isolated from wild-type, heterozygous, and knockout animals. As reported previously (52), two testicular Cdc25C RNA species of 2.1 and 1.9 kb were detected in wild-type animals (Fig. 2A, lane 1). A single species of ∼3.1 kb was detected in the Cdc25C_−/− mice due to insertion of the ∼1.2-kb neomycin phosphotransferase (neo) gene after homologous recombination (lane 3). All three RNA species were present in the testis of heterozygous animals (lane 2). The same blot was stripped and then incubated with probes specific for either neo or GAPDH. The 3.1-kb RNA species detected in testicular RNA derived from heterozygous and null animals was detected with both the m_Cdc25C and neo probes. Results obtained with GAPDH confirm similar amounts of RNA loaded per sample.
FIG. 2.
Northern and Western blot analysis of _Cdc25C_−/− mouse tissues and MEFs. (A) RNA was isolated from the testes of Cdc25C+/+ (WT), Cdc25C+/−(HT), and _Cdc25C_−/− (KO) mice and processed for Northern blotting using probes specific for mCdc25A, mCdc25B, mCdc25C, neomycin phosphotransferase (neo), and GAPDH. (B) Lysates prepared from Sf9 insect cells infected with baculoviruses encoding GST-mCdc25A, GST-mCdc25B, and GST-mCdc25C were lysed and then resolved by SDS-PAGE. Proteins were analyzed by Western blotting using peptide antibody specific for 14 amino acids in the C terminus of mouse Cdc25C (Cdc25C) or with antibody specific for GST. (C) MEFs derived from Cdc25C+/+ (WT) and _Cdc25C_−/− (KO) mouse embryos were incubated with [35S]methionine-cysteine. Cells were lysed, and lysates were incubated with peptide antibody specific for Cdc25C in either the absence (−) or presence (+) of the immunizing peptide. Immunoprecipitates were boiled, and mCdc25C was reimmunoprecipitated prior to SDS-PAGE. Radiolabeled mCdc25C was visualized by fluorography.
Next, fibroblasts were derived from wild-type and knockout MEFs to monitor for the presence of Cdc25C protein. MEFs that had been incubated with [35S]methionine-cysteine were lysed, and lysates were incubated with peptide antibody that specifically recognized mCdc25C but not mCdc25A or mCdc25B (Fig. 2B). Immunoprecipitates were resolved by SDS-PAGE, and the presence of Cdc25C was analyzed by fluorography. A 54-kDa protein, the expected size of mCdc25C, was readily detected in MEFs derived from Cdc25C+/+ animals (Fig. 2C, lane 1), but not in MEFs derived from m_Cdc25C_−/− animals (lane 3). The 54-kDa protein was specifically detected with two independent antibodies, a peptide antibody specific for the C terminus of mCdc25C (Fig. 2C) and an affinity-purified antibody raised against bacterially produced GST-mCdc25C (data not shown). In addition, immunoprecipitation of the 54-kDa protein was specifically blocked by incubation with the immunizing peptide (Fig. 2C, lane 2). These results confirm that the targeted disruption of Cdc25C was successful in producing a null allele of the locus.
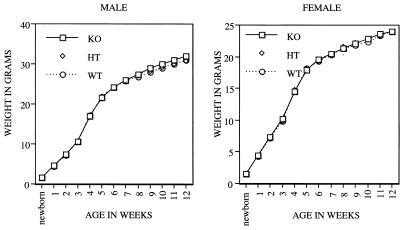
The growth rate of individual mice of all three genotypes from birth to 3 months of age was also determined. As shown in Fig. 3, there was no detectable difference in growth rates between female and male _Cdc25C_−/− mice relative to their wild-type littermates. In addition, kidney, heart, lung, spleen, liver, thymus, testis, and ovary from knockout animals were examined histologically after hematoxylin-eosin staining and appeared normal (data not shown). Cdc25B is expressed to various degrees throughout development, including preimplantation development, and Cdc25A mRNA is first detected at the blastocyst stage and is then expressed ubiquitously throughout the rest of embryonic development. In addition, maternal stores of Cdc25A protein may function to sustain the preimplantation embryo (51, 52). Thus, the early embryonic development of mice lacking Cdc25C may be sustained by the activities of Cdc25A or Cdc25B. Alternatively, other pathways may compensate for the lack of Cdc25C.
FIG. 3.
Growth curves. Cdc25C+/+(WT), Cdc25C+/−(HT), and _Cdc25C_−/−(KO) male (left) and female (right) mice were weighed beginning at birth and at weekly intervals up to 12 weeks. Weights (in grams) represent averages of at least 30 mice per genotype per time point.
Fertility is unaltered in _Cdc25C_−/− mice.
In adult mice, Cdc25C is expressed at highest levels in tissues with mitotically or meiotically active cells, including testis, thymus, ovary, spleen, and intestine (37, 52). Cdc25B expression has been reported to be restricted to the mitotically quiescent somatic cells of the testis, suggesting that it does not contribute to spermatogenesis (52). However, Cdc25A is expressed at high levels in the testis of adult mice, particularly in meiotic germ cells, but it is also present in mitotic germ cells (51, 52). Cdc25A is also expressed in ovary during most stages of follicular development, and Cdc25B expression is detected in the germ line of the ovary (51).
To evaluate testicular and ovarian function, we determined whether fertility was impaired in _Cdc25C_−/− mice. Homozygous crosses demonstrated that both male and female mice with a disruption of Cdc25C were fertile (data not shown). To determine if the fertility of male _Cdc25C_−/− animals could be accounted for by upregulation of either Cdc25A or Cdc25B, total RNA isolated from the testis of Cdc25C+/+ and _Cdc25C_−/− animals was subjected to Northern blot analysis using probes specific for Cdc25A and Cdc25B. As shown in Fig. 2A, both mRNAs were present in testis from Cdc25C+/+ animals, and neither Cdc25A nor Cdc25B mRNA levels were upregulated in _Cdc25C_−/− mice.
T- and B-cell development and function are normal in _Cdc25C_−/− mice.
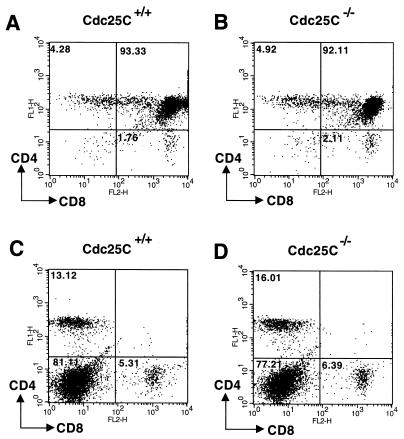
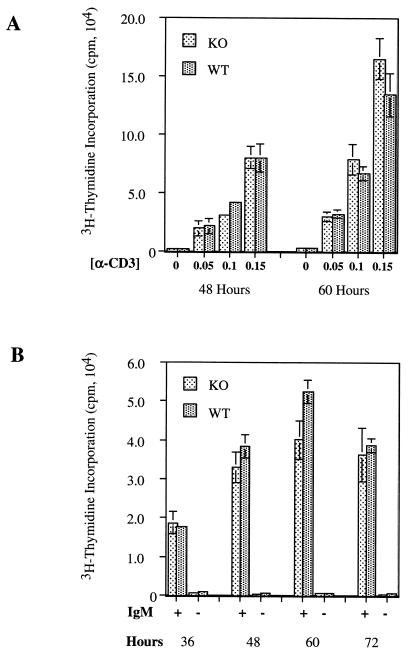
Cdc25C transcripts are also abundant in the thymus and to a lesser extent in spleens of adult mice (37). We therefore examined T- and B-cell development and function in _Cdc25C_−/− mice. FACS analysis using the T-cell markers CD3, CD4, and CD8 revealed the expected number of thymic T cells (data not shown) and demonstrated that thymic T-cell development was normal in _Cdc25C_−/− mice (Fig. 4A and B). In addition, percentages of total single and double positive CD4+ and CD8+ splenic T cells were similar in Cdc25C+/+ and _Cdc25C_−/− mice (Fig. 4C and D). B-cell development appeared normal, based on the presence of similar numbers of B220+ cells in the spleens of Cdc25C+/+ and _Cdc25C_−/− mice (data not shown). The role of Cdc25C in T-cell function was determined by measuring proliferative responses after T-cell receptor cross-linking in vitro. CD4+ splenic T cells isolated from 8-week-old _Cdc25C_−/− and Cdc25C+/+ mice showed similar proliferative responses to anti-CD3 cross-linking in vitro (Fig. 5A). In addition, B-cell proliferative responses to IgM cross-linking were similar in Cdc25C+/+ and _Cdc25C_−/− mice (Fig. 5B). Northern analysis revealed that loss of Cdc25C did not affect mRNA levels of either Cdc25A or Cdc25B in stimulated populations of splenic T and B cells (data not shown).
FIG. 4.
T-cell development is normal in Cdc25C−/− mice. Thymocytes (A and B) and splenocytes (C and D) harvested from 6-week-old mice were stained with FITC-anti-CD4 and PE-anti-CD8. Stained cells were analyzed by flow cytometry gated on lymphocytes, and results are shown as dot plots. The percentage of gated cells in each quadrant is indicated. These data are representative of four independent experiments.
FIG. 5.
T- and B-cell proliferation assays. Splenic CD4+ T cells were stimulated to proliferate by incubation on plate-bound anti-CD3 (A) and B220+ splenocytes were induced to proliferate by incubation with anti-IgM Ab (20 μg/ml) (B) for the indicated times. Cell proliferation was measured by [3H]thymidine incorporation. Standard deviations for triplicate samples are shown as error bars along the y axis. These data are representative of six and eight independent T- and B-cell proliferation assays, respectively.
Cell cycle and checkpoint analysis of cells lacking Cdc25C.
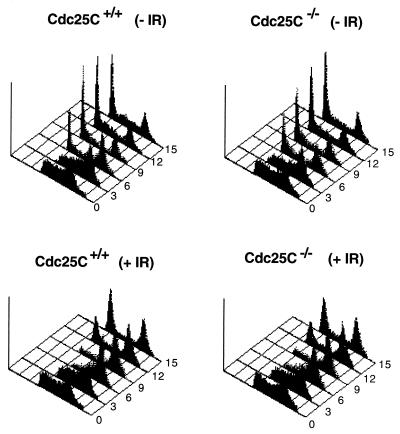
Given that Cdc25C regulates entry into mitosis by dephosphorylating and activating the Cdc2 protein kinase, we next examined the length of time required for cells lacking Cdc25C to traverse from the S-phase of the cell cycle through G2 and into mitosis. Early-passage MEFs were pulse labeled with BrdU to specifically mark cells undergoing DNA replication. Cells were harvested at various times after the incubation and stained with PI and with monoclonal antibodies to BrdU. Processed cells were analyzed for DNA content by FACS. By gating on BrdU-positive cells, cell cycle progression from S through G2/M and into the G1-phase of the cell cycle could be monitored. As shown in Fig. 6A and B and Table 1, the cell cycle profiles obtained for Cdc25C+/+ cells and _Cdc25C_−/− cells were quite similar. By 3 h, 47.39 and 43.56% of BrdU-positive Cdc25C+/+ and _Cdc25C_−/− cells, respectively, had progressed to the G2/M phases of the cell cycle, and by 6 h, 23.15 and 25.09% of BrdU-positive Cdc25C+/+ and _Cdc25C_−/− cells, respectively, had progressed to the G1 phase of the cell cycle. These findings indicate that the time required for cells to progress from the S to the G1 phase of the cell cycle is unaltered in murine cells lacking Cdc25C
FIG. 6.
Cell cycle and checkpoint analysis of cells lacking Cdc25C. MEFs prepared from Cdc25C+/+ and _Cdc25C_−/− mice were pulse labeled with BrdU for 1 h and then treated with 0 (−IR) or 4 Gy (+IR) of γ-irradiation. Cells were harvested at the indicated times (hours) and stained with PI and for BrdU. Cellular DNA content of BrdU-positive cells was analyzed by flow cytometry.
TABLE 1.
Cell cycle and checkpoint analysis of cells lacking Cdc25Ca
| Time (h) | Genotype | % of cells | |||||
|---|---|---|---|---|---|---|---|
| Without ionizing radiation | With ionizing radiation | ||||||
| G1 | S | G2/M | G1 | S | G2/M | ||
| 3 | WT | 1.76 | 51.27 | 47.39 | 0.52 | 57.83 | 41.98 |
| KO | 7.12 | 49.75 | 43.56 | 1.17 | 56.94 | 42.21 | |
| 6 | WT | 23.15 | 31.43 | 45.81 | 0.29 | 36.37 | 63.63 |
| KO | 25.09 | 34.20 | 41.13 | 1.45 | 42.25 | 56.71 | |
| 9 | WT | 38.61 | 27.53 | 34.26 | 6.29 | 27.88 | 66.06 |
| KO | 35.99 | 31.62 | 32.79 | 7.57 | 31.62 | 61.12 | |
| 12 | WT | 40.11 | 32.10 | 28.05 | 24.26 | 26.42 | 49.64 |
| KO | 38.68 | 30.43 | 31.20 | 20.11 | 27.86 | 52.36 | |
| 15 | WT | 37.29 | 33.83 | 29.13 | 36.46 | 22.13 | 41.65 |
| KO | 41.56 | 27.85 | 30.87 | 31.20 | 25.67 | 43.41 |
In humans, Xenopus, and fission yeast, Cdc25C has been shown to be an important target of the G2 DNA damage checkpoint (31, 40, 55, 56). Therefore, we examined murine cells lacking Cdc25C for their ability to delay cell cycle progression in response to DNA damage. BrdU-labeled MEFs were gamma irradiated and monitored for their ability to delay the G2 phase of the cell cycle. As shown in Fig. 6C and D and Table 1, the cellular response to DNA damage was intact in cells lacking Cdc25C. A predominant G1 peak of cells was not observed until 12 h after irradiation for both Cdc25C+/+ and _Cdc25C_−/− cells (Fig. 6C and D). This is in contrast to control cells, where a strong G1 peak was observed by 6 h (Fig. 6A and B). Thus, murine cells lacking Cdc25C are fully capable of mounting a G2 DNA checkpoint response.
Next, the phosphorylation status of Cdc2 was monitored in early-passage fibroblasts derived from wild-type and knockout mouse embryos. The phosphorylation of Cdc2 varies as a function of the cell cycle (1, 45). In G1, Cdc2 is not bound to the B-type cyclins and is unphosphorylated. During S and G2, Cdc2 binds to the B-type cyclins and is subsequently phosphorylated on Thr-161, Thr-14, and Tyr-15. Thr-161 phosphorylation does not alter the electrophoretic mobility of Cdc2. However, the electrophoretic mobility of Cdc2 decreases upon phosphorylation of either Thr-14 or Tyr-15, and a further decrease in mobility is observed when Cdc2 is phosphorylated on both residues simultaneously. Loss of phosphorylation at Thr-14 and Tyr-15 occurs at the onset of mitosis, when Cdc2 is dephosphorylated and activated by the Cdc25C protein phosphatase. Thus, Cdc2 from G1- and M-phase cells migrates with the fastest electrophoretic mobility on SDS gels, whereas the two slower electrophoretic forms of Cdc2 are observed in the S and G2 phases of the cell cycle. Therefore, changes in the electrophoretic mobility of Cdc2 can be used as a specific indicator both of Thr-14 and Tyr-15 phosphorylation and cell cycle position (S and G2).
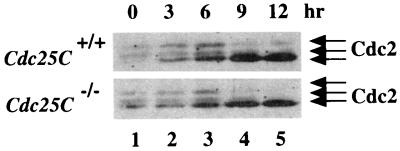
Cells were arrested in early S phase by first culturing in medium containing low serum and then releasing into complete medium containing aphidicolin. Cells were harvested prior to release (time 0) or at various times after release from the block and analyzed for DNA content by FACS (data not shown) or processed to monitor for Cdc2 phosphorylation status (Fig. 7). Cdc25C+/+ and _Cdc25C_−/− cells that escaped the block were in S phase by 3 h after release, in G2 phase by 6 h after release, and in the M and G1 phases of the cell cycle by 9 and 12 h after release. Lysates prepared from cells at each time point were incubated with p13_suc1_-agarose, and precipitates were resolved by SDS-PAGE and probed with Cdc2-specific antibody. As shown in Fig. 7, Cdc2 existed in three electrophoretic forms in early S (lane 1)-, S (lane 2)-, and G2 (lane 3)-phase cells. As expected, a loss of the two slower electrophoretic forms of Cdc2 with a concomitant increase in the fastest electrophoretic form of Cdc2 was observed as cells entered mitosis (lane 4) and moved into G1 (lane 5). We did not observe any difference in cell cycle progression or Cdc2 phosphorylation status in Cdc25C+/+ and _Cdc25C_−/− cells.
FIG. 7.
Phosphorylation status of Cdc2 in cells lacking Cdc25C. Early-passage MEFs prepared from Cdc25C+/+ and _Cdc25C_−/− mice were synchronized in early S phase. Cells were harvested prior to release (time 0) or at 3, 6, 9, or 12 h after release. Cell lysates were prepared and incubated with p13_suc1_-agarose to precipitate Cdc2. Precipitates were resolved on a 10% SDS gel, and Cdc2 was detected by Western blotting. The arrows indicate the three electrophoretic forms of Cdc2.
Concluding remarks.
Mice lacking Cdc25C were viable, developed normally, and did not display any obvious abnormalities. It is unknown at this time whether Cdc25A and/or Cdc25B compensates for lack of Cdc25C throughout mouse development or whether other compensatory mechanisms exist in mice lacking Cdc25C. Analysis of Cdc25A and Cdc25B singly deficient mice along with doubly deficient mice may elucidate the contribution made by individual members of the Cdc25 family of protein phosphatases to cell cycle regulation throughout development and in adult mouse tissues.
ACKNOWLEDGMENTS
We thank Peter Donovan for providing peptide antibody specific for mCdc25C. We thank the Siteman Cancer Center ES stem cell core for electroporation of the targeting construct into RW4 cells and Mike White for the blastocyst injections. We thank B. Sleckman and A. Chan for helpful suggestions and comments. We thank Y. Zhao and L. He for technical support.
This work was supported in part by a scholarship from the Lucille P. Markey foundation to J.H. M.-S.C. is an Associate and H.P.-W. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Atherton-Fessler S, Liu F, Gabrielli B, Lee M S, Peng C-Y, Piwnica-Worms H. Cell cycle regulation of the p34cdc2 inhibitory kinases. Mol Biol Cell. 1994;5:989–1001. doi: 10.1091/mbc.5.9.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blomberg I, Hoffman I. Ectopic expression of Cdc25A accelerates the G1/S transition and leads to premature activation of cyclin E- and cyclin A-dependent kinases. Mol Cell Biol. 1999;19:6183–6194. doi: 10.1128/mcb.19.9.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chamberlain J P. Fluorographic detection of radioactivity in polyacrylamide gels with the water soluble fluor sodium salicylate. Anal Biochem. 1979;98:132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- 4.Chauhan S, Gottesman M. Construction of a new universal vector for insertional mutagenesis by homologous recombination. Gene. 1992;120:281–286. doi: 10.1016/0378-1119(92)90106-y. [DOI] [PubMed] [Google Scholar]
- 5.Conklin D S, Galaktionov K, Beach D. 14-3-3 proteins associate with cdc25 phosphatases. Proc Natl Acad Sci USA. 1995;92:7892–7896. doi: 10.1073/pnas.92.17.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalal S N, Schweitzer C M, Gan J, Decaprio J. Cytoplasmic localization of human Cdc25C during interphase requires an intact 14-3-3 binding site. Mol Cell Biol. 1999;19:4465–4479. doi: 10.1128/mcb.19.6.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davezac N, Baldin V, Gabrielli B, Forrest A, Theis-Febvre N, Yashida M, Ducommun B. Regulation of CDC25B phosphatases subcellular localization. Oncogene. 2000;19:2179–2185. doi: 10.1038/sj.onc.1203545. [DOI] [PubMed] [Google Scholar]
- 8.Ducommun B, Draetta G, Young P, Beach D. Fission yeast cdc25 is a cell cycle regulated protein. Biochem Biophys Res Commun. 1990;167:301–309. doi: 10.1016/0006-291x(90)91765-k. [DOI] [PubMed] [Google Scholar]
- 9.Dunphy W G, Kumagai A. The cdc25 protein contains an intrinsic phosphatase activity. Cell. 1991;67:189–196. doi: 10.1016/0092-8674(91)90582-j. [DOI] [PubMed] [Google Scholar]
- 10.Fantes P. Epistatic gene interactions in the control of division in fission yeast. Nature. 1979;279:428–430. doi: 10.1038/279428a0. [DOI] [PubMed] [Google Scholar]
- 11.Gabrielli B G, Clark J M, McCormack A K, Ellem K A. Hyperphosphorylation of the N-terminal domain of Cdc25 regulates activity toward cyclin B1/Cdc2 but not cyclin A/Cdk2. J Biol Chem. 1997;272:28607–28614. doi: 10.1074/jbc.272.45.28607. [DOI] [PubMed] [Google Scholar]
- 12.Gabrielli B G, De Souza C P C, Tonks I D, Clark J M, Hayward N K. Cytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cells. J Cell Sci. 1996;109:1081–1093. doi: 10.1242/jcs.109.5.1081. [DOI] [PubMed] [Google Scholar]
- 13.Galaktionov K, Beach D. Specific activation of cdc25 tyrosine phosphatases by B-type cyclins: evidence for multiple roles of mitotic cyclins. Cell. 1991;67:1181–1194. doi: 10.1016/0092-8674(91)90294-9. [DOI] [PubMed] [Google Scholar]
- 14.Galaktionov K, Lee A K, Eckstein J, Draetta G, Meckler J, Loda M, Beach D. CDC25 phosphatases as potential human oncogenes. Science. 1995;269:1575–1577. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 15.Gasparotto D, Maestro R, Piccinin S, Vukosavljevic T, Barzan L, Sulfaro S, Boiocchi M. Overexpression of CDC25A and CDC25B in head and neck cancers. Cancer Res. 1997;57:2366–2368. [PubMed] [Google Scholar]
- 16.Gautier J, Solomon M J, Booher R N, Bazan J F, Kirschner M W. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- 17.Graves P R, Lovly C M, Uy G L, Piwnica-Worms H. Localization of human Cdc25C is regulated both by nuclear export and 14-3-3 binding. 2001. Oncogene, in press. [DOI] [PubMed] [Google Scholar]
- 18.Graves P R, Yu L, Schwarz J K, Gales J, Sausville E A, O'Connor P M, Piwnica-Worms H. The Chk1 protein kinase and the Cdc25C regulatory pathway are targets of the anticancer agent UCN-01. J Biol Chem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez S, Hernandez L, Bea S, Pinyol M, Nayach I, Bellosillo B, Nadal A, Ferrer A, Fernandez P L, Montserrat E, Cardesa A, Campo E. Cdc25A and the splicing variant cdc25B2, but not cdc25B1, -B3 or -C, are over-expressed in aggressive human non-Hodgkin's lymphomas. Int J Cancer. 2000;89:148–152. [PubMed] [Google Scholar]
- 20.Hoffmann I, Clarke P R, Marcote M J, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2-cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann I, Draetta G, Karsenti E. Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E dependent phosphorylation at the G1/S transition. EMBO J. 1994;13:4302–4310. doi: 10.1002/j.1460-2075.1994.tb06750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honda R, Ohba Y, Nagata A, Okayama H, Yasuda H. Dephosphorylation of human p34cdc2 kinase on both Thr-14 and Tyr-15 by human cdc25B phosphatase. FEBS Lett. 1993;318:331–334. doi: 10.1016/0014-5793(93)80540-b. [DOI] [PubMed] [Google Scholar]
- 23.Iavarone A, Massague J. Repression of the CDK activator Cdc25A and cell-cycle arrest by the cytokine TGF-β in cells lacking the CDK inhibitor p15. Nature. 1997;387:417–422. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- 24.Izumi T, Maller J L. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol Biol Cell. 1993;4:1337–1350. doi: 10.1091/mbc.4.12.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izumi T, Walker D H, Maller J M. Periodic changes in phosphorylation of the Xenopus cdc25 phosphatase regulate its activity. Mol Biol Cell. 1992;3:927–939. doi: 10.1091/mbc.3.8.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jinno S, Suto K, Nagata A, Igarashi M, Kanaoka Y, Nojima H, Okayama H. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J. 1994;13:1549–1556. doi: 10.1002/j.1460-2075.1994.tb06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kakizuka A, Sebastian B, Borgmeyer U, Hermans-Borgmeyer I, Bolado J, Hunter T, Hoekstra M F, Evans R M. A mouse cdc25 homolog is differentially and developmentally expressed. Genes Dev. 1992;6:578–590. doi: 10.1101/gad.6.4.578. [DOI] [PubMed] [Google Scholar]
- 28.Kuang J, Ashorn C L, Gonzalez-Kuyvenhoven M, Penkala J E. cdc25 is one of the MPM-2 antigens involved in the activation of maturation-promoting factor. Mol Biol Cell. 1994;5:135–145. doi: 10.1091/mbc.5.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumagai A, Dunphy W G. Purification and molecular cloning of Plx1, a cdc25-regulatory kinase from Xenopus egg extracts. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- 30.Kumagai A, Dunphy W G. Regulation of the cdc25 protein during the cell cycle in Xenopus extracts. Cell. 1992;70:139–151. doi: 10.1016/0092-8674(92)90540-s. [DOI] [PubMed] [Google Scholar]
- 31.Kumagai A, Yakowec P S, Dunphy W G. 14-3-3 proteins act as negative regulators of the mitotic inducer Cdc25 in Xenopus egg extracts. Mol Biol Cell. 1998;9:345–354. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lammer C, Wagerer S, Saffrich R, Mertens D, Ansorge W, Hoffmann I. The cdc25B phosphatase is essential for the G2/M phase transition in human cells. J Cell Sci. 1998;111:2445–2453. doi: 10.1242/jcs.111.16.2445. [DOI] [PubMed] [Google Scholar]
- 33.Lee M S, Ogg S, Xu M, Parker L L, Donoghue D J, Maller J L, Piwnica-Worms H. cdc25+ encodes a protein phosphatase that dephosphorylates p34cdc2. Mol Biol Cell. 1992;3:73–84. doi: 10.1091/mbc.3.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mailand N, Falck J, Lukas C, Syljuasen R G, Welcker M, Bartek J, Lukas J. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 35.Millar J B A, Blevitt J, Gerace L, Sadhu K, Featherstone C, Russell P. p55cdc25 is a nuclear protein required for the initiation of mitosis in human cells. Proc Natl Acad Sci USA. 1991;88:10500–10504. doi: 10.1073/pnas.88.23.10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagata A, Igarashi M, Jinno S, Suto K, Okayama H. An additional homolog of the fission yeast cdc25+ gene occurs in humans and is highly expressed in some cancer cells. New Biol. 1991;3:959–968. [PubMed] [Google Scholar]
- 37.Nargi J L, Woodford-Thomas T A. Cloning and characterization of a cdc25 phosphatase from mouse lymphocytes. Immunogenetics. 1994;39:99–108. doi: 10.1007/BF00188612. [DOI] [PubMed] [Google Scholar]
- 38.Nishijima H, Nishitani H, Seki T, Nishimoto T. A dual-specificity phosphatase Cdc25B is an unstable protein and triggers p34(cdc2)/cyclin B activation in hamster BHK21 cells arrested with hydroxyurea. J Cell Biol. 1997;138:1105–1116. doi: 10.1083/jcb.138.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker L L, Atherton-Fessler S, Lee M S, Ogg S, Falk F L, Swenson K I, Piwnica-Worms H. Cyclin promotes the tyrosine phosphorylation of p34cdc2 in a wee1+ dependent manner. EMBO J. 1991;10:1255–1263. doi: 10.1002/j.1460-2075.1991.tb08067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng C-Y, Graves P R, Thoma R S, Wu Z, Shaw A, Piwnica-Worms H. Mitotic- and G2-checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine 216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 41.Piwnica-Worms H. Expression of proteins in insect cells using baculovirus vectors. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 42.Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45:145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- 43.Sebastian B, Kakizuka A, Hunter T. Cdc25M2 activation of cyclin-dependent kinases by dephosphorylation of threonine-14 and tyrosine-15. Proc Natl Acad Sci USA. 1993;90:3521–3524. doi: 10.1073/pnas.90.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sexl V, Diehl J, Sherr C, Ashmun R, Beach D, Roussel M F. A rate limiting function of cdc25A for S phase entry inversely correlates with tyrosine dephosphorylation of Cdk2. Oncogene. 1999;18:573–582. doi: 10.1038/sj.onc.1202362. [DOI] [PubMed] [Google Scholar]
- 45.Solomon M J, Lee T, Kirschner M W. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol Biol Cell. 1992;3:13–27. doi: 10.1091/mbc.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strausfeld U, Fernandez A, Capony J-P, Girard F, Lautredou N, Derancourt J, Labbe J-C, Lamb N J C. Activation of p34cdc2 protein kinase by microinjection of human cdc25C into mammalian cells. J Biol Chem. 1994;269:5989–6000. [PubMed] [Google Scholar]
- 47.Strausfeld U, Labbe J C, Fesquet D, Cavadore J C, Picard A, Sadhu K, Russell P, Doree M. Dephosphorylation and activation of a p34cdc2/cyclin B complex in vitro by human cdc25 protein. Nature. 1991;351:242–245. doi: 10.1038/351242a0. [DOI] [PubMed] [Google Scholar]
- 48.Takemasa I, Yamamoto H, Sekimoto M, Ohue M, Noura S, Miyake Y, Matsumoto T, Aihara T, Tomita N, Tamaki Y, Sakita I, Kikkawa N, Matsuura N, Shiozaki H, Monden M. Overexpression of CDC25B phosphatase as a novel marker of poor prognosis of human colorectal carcinoma. Cancer Res. 2000;60:3043–3050. [PubMed] [Google Scholar]
- 49.Terada Y, Tatsuka M, Jinno S, Okayama H. Requirement for tyrosine phosphorylation of Cdk4 in G1 arrest induced by ultraviolet irradiation. Nature. 1995;376:358–362. doi: 10.1038/376358a0. [DOI] [PubMed] [Google Scholar]
- 50.Villa-Moruzzi E. Activation of the cdc25C phosphatase in mitotic HeLa cells. Biochem Biophys Res Commun. 1993;196:1248–1254. doi: 10.1006/bbrc.1993.2386. [DOI] [PubMed] [Google Scholar]
- 51.Wickramasinghe D, Becker S, Ernst M K, Resnick J L, Centanni J M, Tessarollo L, Grabel L B, Donovan P J. Two CDC25 homologues are differentially expressed during mouse development. Development. 1995;121:2047–2056. doi: 10.1242/dev.121.7.2047. [DOI] [PubMed] [Google Scholar]
- 52.Wu S, Wolgemuth D J. The distinct and developmentally regulated patterns of expression of members of the mouse Cdc25 gene family suggest differential functions during gametogenesis. Dev Biol. 1995;170:195–206. doi: 10.1006/dbio.1995.1207. [DOI] [PubMed] [Google Scholar]
- 53.Wu W, Fan Y-H, Kemp B L, Walsh G, Mao L. Overexpression of cdc25A and cdc25B is frequent in primary non-small cell lung cancer but is not associated with overexpression of c-myc. Cancer Res. 1998;58:4082–4085. [PubMed] [Google Scholar]
- 54.Yao Y, Slosberg E D, Wang L, Hibshoosh H, Zhang U-J, Xing W-Q, Santella R M, Weinstein I B. Increased susceptibility to carcinogen-induced mammary tumors in MMTV-Cdc25B transgenic mice. Oncogene. 1999;18:5159–5166. doi: 10.1038/sj.onc.1202908. [DOI] [PubMed] [Google Scholar]
- 55.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint in fission yeast requires Cdc25p phosphorylation by Cds1p or Chk1p. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- 56.Zeng Y, Piwnica-Worms H. The DNA damage and replication checkpoints in fission yeast require nuclear exclusion of the Cdc25 phosphatase via 14-3-3 binding. Mol Cell Biol. 1999;19:7410–7419. doi: 10.1128/mcb.19.11.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]