Two-Component Signal Transduction in Bacillus subtilis: How One Organism Sees Its World (original) (raw)
Variability and adaptability are crucial characteristics of organisms possessing the ability to survive and prosper in a wide variety of environmental conditions. The most adaptable bacteria contain a large reservoir of genetic information encoding biochemical pathways designed to cope with a variety of environmental situations. Organisms that have the genetic capability to respond to altered conditions do so when stimulated by specific signals. Recognition of specific signals and conversion of this information into specific transcriptional or behavioral responses is the essence of signal transduction.
A mechanism commonly found in bacteria for signal transduction is the two-component system (23, 26). Its basis is the conversion of signal recognition to a chemical entity, i.e., a phosphoryl group, that modifies the functional activity of proteins. Signal recognition and transduction are the province of the sensor histidine kinase component of the system. This protein has separable sensor and histidine phosphotransferase domains that function to recognize (bind) the signal, causing the kinase to autophosphorylate a histidine residue of the phosphotransferase domain (Fig. 1). The phosphoryl group is subsequently transferred to the second component protein, the response regulator, where it resides as an acyl phosphate of an aspartic acid residue. The response regulator consists of the phosphorylatable aspartate domain and an output domain that is activated to carry out its function by conformational or, perhaps, electrostatic alterations induced by the phosphoryl group. In most cases, the response regulator is a transcription activator for genes whose products are specifically utilized to respond to the unique nature of a given input signal. In the chemotaxis system of bacteria, the response regulator determines the direction of rotation of the flagellar motor. The basics of the signal transduction mechanism remain the same regardless of the input signal or the function of the response regulator.
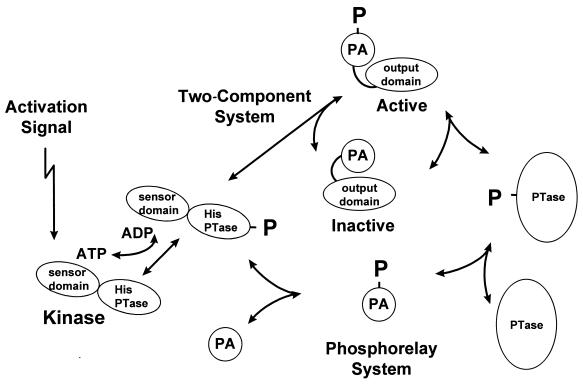
FIG. 1.
Schematic view of two-component and phosphorelay systems. Activation signals recognized by sensor domains of histidine kinases result in autophosphorylation of a histidine in the histidine phosphotransferase domain (His PTase). The phosphoryl group (P) is transferred directly to the phosphorylated aspartate domain (PA) of a response regulator in a two-component system, causing a conformational change that activates the output domain. In a phosphorelay, the phosphoryl group is transferred to a PA domain that serves as a substrate for a phosphotransferase whose role is to transfer the phosphoryl group to the PA domain of a response regulator. Note that all the steps are reversible in many systems, which may result in dephosphorylation in the absence of a signal.
This phosphoryl group-based signal transduction mechanism exists in two major conformations in microorganisms: the two-component system and a four-component system termed the phosphorelay (Fig. 1). Signal interpretation and transduction by histidine kinases are the same in both, but the target of the kinase in a phosphorelay is a single-domain response regulator consisting of only the phosphorylated aspartate domain. This phosphorylated protein serves as a substrate for a phosphotransferase that transfers the phosphoryl group to a response regulator-transcription factor. The phosphotransferase is transiently phosphorylated on a histidine during this process. In a phosphorelay, the phosphoryl group is transferred in the order His-Asp-His-Asp, which differs from the His-Asp series of a two-component system. In the first-discovered phosphorelay used to initiate sporulation in Bacillus subtilis, all of the components (domains) reside on different proteins (4). Subsequently discovered phosphorelays in bacteria, fungi, and plants use composite proteins where the kinase and first response regulator domain and sometimes the phosphotransferase domain are contiguous within a single polypeptide chain (1, 6). The sporulation initiation phosphorelay is a signal integration circuit that processes both positive and negative signals, which suggests that phosphorelays are used where a number of opposing signals must be interpreted by the signal transduction system (22).
GENOME ANALYSIS OF TWO-COMPONENT SYSTEMS
Knowing the complete sequence of the B. subtilis chromosome allowed analysis of the number and kinds of two-component systems in this organism (14). The structural and functional principles for these analyses were the conserved ATP-binding site characteristic of sensor histidine kinases in conjunction with a conserved histidine motif and the overall similarity of the phosphorylated aspartate domains of response regulators (23). Using these criteria, 36 histidine kinases and 34 response regulators were found among the open reading frames identified in the genome (see Table 1). Comparing the kinases found to those of the distantly related gram-negative microorganism Escherichia coli revealed that none of the B. subtilis enzymes were composite kinases in which a phosphorylatable response regulator domain was contiguous with the kinase polypeptide. E. coli has five of these composite kinases that are believed to function in phosphorelays similar to the sporulation phosphorelay (19, 20).
TABLE 1.
Two-component systems in B. subtilis
| Classa | Familyb | Kinase | Response regulator | Organizationc | Adaptive role |
|---|---|---|---|---|---|
| I | Others-B | LytS | LytT | HR | Rate of autolysis |
| YesM | YesN | HR | Unknown and nonessential | ||
| YwpD | Orphand | Unknown and nonessential | |||
| II | NarL | DegS | DegU | HR | Degradative enzyme and competence |
| YhcY | YhcZ | HR | Unknown and nonessential | ||
| YvqE | YvqC | HR | Unknown and nonessential | ||
| YvfT | YvfU | HR | Unknown and nonessential | ||
| YocF | YocG | HR | Unknown and nonessential | ||
| YdfH | YdfI | HR | Unknown and nonessential | ||
| YxjM | YxjL | HR | Unknown and nonessential | ||
| YfiJ | YfiK | HR | Unknown and nonessential | ||
| ComP | ComA | HR | Early competence | ||
| IIIA | OmpR | YcbM | YcbL | RH | Unknown and nonessential |
| YxdK | YxdJ | RH | Unknown and nonessential | ||
| YtsB | YtsA | RH | Unknown and nonessential | ||
| YvcQ | YvcP | RH | Unknown and nonessential | ||
| YbdK | YbdJ | RH | Unknown and nonessential | ||
| YvqB | YvqA | RH | Unknown and nonessential | ||
| YkoH | YkoG | RH | Unknown and nonessential | ||
| YrkQ | YrkP | RH | Unknown and nonessential | ||
| YclK | YclJ | RH | Unknown and nonessential | ||
| YvrG | YvrH | RH | Unknown and nonessential | ||
| YycG | YycF | RH | Essential functions | ||
| ResE | ResD | RH | Aerobic and anaerobic respiration | ||
| PhoR | PhoP | RH | Alkaline phosphatase and phosphodiesterase | ||
| YccG | YccH | RH | Unknown and nonessential | ||
| IIIB | NtrB | KinA | Spo0F | Orphan | Initiation of sporulation |
| KinB | Spo0F | Orphan | Initiation of sporulation | ||
| KinC | Orphan | ||||
| YkvD | Orphan | ||||
| YkrQ | Orphan | ||||
| IV | Others-A | YufL | YufM | HR | Unknown and nonessential |
| YdbF | YdbG | HR | Unknown and nonessential | ||
| CitS | CitT | HR | Mg2+/citrate transport | ||
| YcbA | YcbB | HR | Unknown and nonessential | ||
| V | CheY | CheA | CheY | RH | Chemotaxis |
The CheY protein is the single example in E. coli of a response regulator consisting of only the phosphorylatable aspartate domain. Three of these were found in B. subtilis. These include a close homologue of CheY as well as Spo0F, of the sporulation phosphorelay, and YneI, a protein of unknown function. While most response regulators are transcription factors dependent on phosphorylation for activity, phosphorylation of these single-domain response regulators serves other functions, such as interaction with the flagellar motor in the case of CheY, or as phosphointermediates in the phosphorelay in the case of Spo0F.
CLASSIFICATION OF HISTIDINE KINASES AND RESPONSE REGULATORS
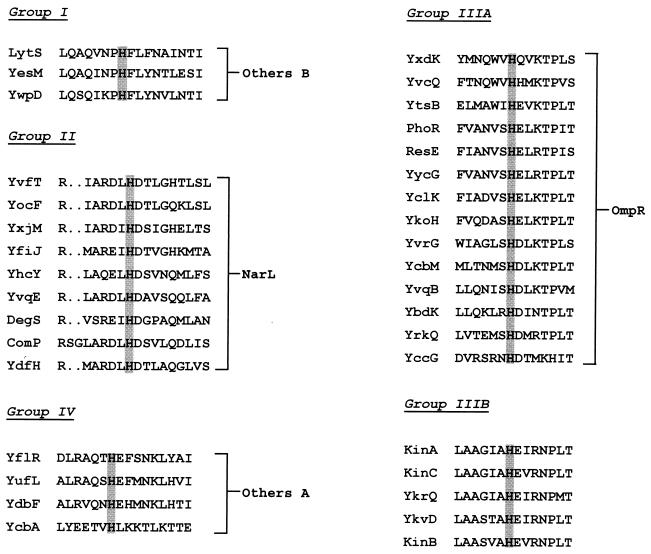
The classification of histidine kinases and response regulators into related groups was not accomplished on the basis of overall protein homology. The sensor domains of histidine kinases differ greatly, possibly reflecting the diversity of molecules sensed by the microorganism. Histidine kinases are characterized by the presence of a conserved ATP-binding site that, while it distinguishes them from other proteins, does little to differentiate them. Examination of the region around the histidine that becomes phosphorylated was more informative. The histidine motifs fell into five homology classes of which two, IIIA and IIIB, were closely related (Fig. 2).
FIG. 2.
Classification of kinases by the sequence around the phosphorylated histidine. Kinases were sorted into classes on the basis of the sequence relationships of the residues on either side of the phosphorylated histidine. The classes were related to their response regulator based on the homology of the response regulator output domain to those of E. coli (19). Note the orphan kinases of group IIIB were related to NtrB of E. coli through the homology of the residues surrounding the histidine to NtrB, not through homologies to NtrC, a response regulator which does not exist in B. subtilis. Gaps introduced to maximize alignment are indicated by the dots.
Response regulators present a special problem in classification since the phosphorylated aspartate domain is highly conserved in them. This suggests the observed conservation of amino acids occurs because of structural similarities of this domain. High-resolution structural studies of the CheY response regulator of E. coli and Spo0F of B. subtilis showed a remarkable similarity in structure between these two molecules. Despite the conservation of amino acids, alanine-scanning mutagenesis studies of Spo0F revealed that only a small number of residues around the active site determine specificity of interaction with other components of the signaling pathway (29). Thus, amino acid similarity per se is a valid criterion for functional relatedness but does not allow distinction among response regulators.
Most of the response regulators could be classified by the relatedness of their output domains. Structural determinations of this domain of the E. coli OmpR and NarL response regulators provided a basis for relating similarity to structure. Alignment of the C-terminal domains of B. subtilis response regulators with the amino acid sequence of the OmpR DNA-binding domain revealed a group of response regulators with high homology to E. coli OmpR (Fig. 3). The most informative conserved amino acids were the residues making up the hydrophobic core of this domain (17). All of the response regulators falling in this group were paired with a kinase classified as group IIIA by the homology around the histidine residue. One exception to this rule is YccH, which has weak similarity to OmpR (Fig. 3). A similar study using the E. coli NarL output domain identified nine response regulators with high homology for those residues required for proper folding of the domain (2) (Fig. 4). Interestingly, all of these response regulators are paired with a kinase of group II. None of the kinases from group II or IIIA were paired with a response regulator of a different type with the possible exception of YccG. Using this method of analysis, 23 of the response regulators were found to be related to either OmpR or NarL. Comparison of the entire catalytic domain of the kinases to E. coli kinases revealed that class II kinases were most related to NarX homologues and class IIIA kinases were most related to EnvZ homologues as expected (data not shown). Since classification of the kinases was based on homology around the phosphorylated histidine and this region most certainly interacts with the active-site region of the phosphorylated aspartate domain of response regulators, the simplest conclusion for the observed relationships is that the catalytic domain of the kinase and both domains of the response regulator evolved as a unit from a common ancestor. Consistent with this conclusion is the observation that gene order in the transcription unit in which they reside is preserved within classes (see Table 1). The origins of the diverse sensor domains of the kinases remain to be uncovered, but clear subgroups exist within each group with sensor domains of similar size and membrane configuration. Some of the kinases within subgroups clearly evolved from a common progenitor (e.g., PhoR and ResE).
FIG. 3.
Relationships of response regulators to the output domain of OmpR. Amino acid sequences of response regulators were compared to the sequence of the output domain of OmpR of E. coli. The shaded residues are the residues making up the hydrophobic core of the domain (17). Gaps introduced to maximize alignment are indicated by the dashes.
FIG. 4.
Relationships of response regulators to the output domain of NarL. Amino acid sequences of response regulators were compared to the sequence of the output domain of NarL of E. coli. The shaded residues are crucial for correct folding of the domain (2). Gaps introduced to maximize alignment are indicated by the dashes.
Three kinase-response regulator pairs were classified in group I, and they were related to the Others B group of E. coli (19). Similarly, four pairs were classified in group IV and were related to the Others A group of E. coli. CheY and CheA are alone in their classification. YneI is an orphan single-domain response regulator for which there is no basis for classification. Finally, at the level of the kinases, the C-terminal domains of YccG, YbdK, and YcbA do not align perfectly with the other members of their class.
ORPHAN KINASES OF CLASS IIIB
Five kinases originally grouped into class IIIB by their relatedness around the phosphorylated histidine were present on the chromosome without a linked response regulator gene. Two of these orphan kinases, KinA and KinB, are the major kinases responsible for phosphate input into the phosphorelay initiating sporulation (28). KinC was identified as a kinase that phosphorylates mutant forms of the Spo0A response regulator allowing bypass of the phosphorelay in sporulation (13, 15). YkvD is known to phosphorylate certain Spo0F mutants and bypass both KinA and KinB (12). YkrQ has not been implicated in the phosphorelay. These five kinases have residues around the phosphorylated histidine with sequence homology to those found in the NtrB kinase of E. coli. There is no formal equivalent to the NtrB-NtrC two-component system in B. subtilis, and the regulation of nitrogen metabolism is very different in the two organisms. In addition, none of the B. subtilis response regulators contain an NtrC-like ATPase domain required for ς54 activity despite the presence of ς54 and genes transcribed by it. The orphan kinases of class IIIB have no known relationship to nitrogen metabolism, and their sequence similarity around the phosphorylated histidine suggests they may all act as transducers of different signals in sporulation (11).
REGULATORY FUNCTIONS OF TWO-COMPONENT SYSTEMS
Several two-component systems have been extensively studied in B. subtilis and the genes they regulate are known. They include such systems as CheA-CheY in chemotaxis (24), PhoR-PhoP in phosphate regulation (27), ResE-ResD in anaerobic gene activation (21), ComP-ComA in competence (9), and DegS-DegU in degradative enzyme regulation (5). The CitS-CitT system may be involved in Mg2+/citrate transport based on its close similarity to a system in E. coli, and LytS-LytT may be involved in autolysis regulation based on similarity to a system in Staphylococcus aureus (Table 1). The remainder of the systems identified from genome analysis have so little similarity to characterized systems from other organisms that a tentative functional assignment is unwarranted.
In a directed gene knockout study of the response regulators of the unknown two-component systems shown in Table 1, only the YycG-YycF system was found to be essential for growth (7). The other response regulator null mutations did not noticeably affect colony morphology, growth, or sporulation on laboratory media. It is probably safe to conclude that most two-component regulation is used for enhancing the versatility of the response of the organism to environmental stimuli by the regulation of normally unexpressed genes.
It was somewhat surprising that so few of the kinases were related to those of E. coli by similarities in sequence of their sensor domains. This likely reflects the different environments the two organisms occupy and, therefore, the different signals they must process. Spore-forming B. subtilis might be caught dead in an intestine, but, unlike E. coli, would not grow there. On the other hand, a haystack is loaded with B. subtilis and probably contains E. coli only if a cow happened to stop for a bite.
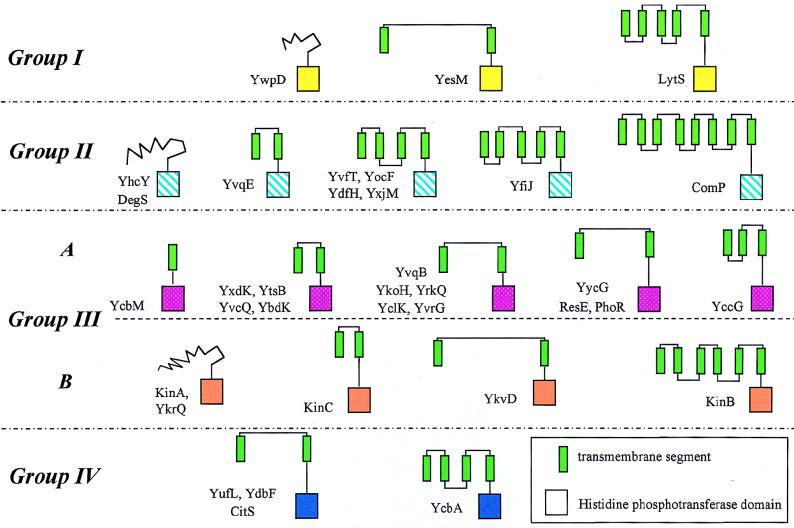
The kinases, with the exception of five, are believed to be embedded in the cellular membrane based on computer identification of transmembrane domains. Some of the kinases have large periplasmic domains, whereas, in others, the sensor domains are mostly hydrophobic membrane domains. There exists a wide diversity of types of sensor domains (Fig. 5). Some of these may be ligand binding, and others, such as that of KinB, are most consistent with a transport role. In view of their diversity and the nonspecific homology of amino acids making up transmembrane domains, the evolutionary relationships between sensor domains is subject to uncertainty and, therefore, is best left uninterpreted.
FIG. 5.
Schematic structures of the kinases. Groups were determined from the homology of the residues surrounding the phosphorylated histidine of the histidine phosphotransferase domain. Related domains are the same color, and green rectangles are likely transmembrane segments.
CYTOPLASMIC LINKERS BETWEEN SENSOR AND HISTIDINE PHOSPHOTRANSFERASE DOMAINS
The sensor domains of membrane kinases are connected to the histidine phosphotransferase domains through a cytoplasmic linker that starts at the end of the last membrane-spanning domain and ends at the phosphorylated histidine motif. These linkers are of variable size but roughly fall into three length classes: ≤40, 60 to 80, and 130 to 170 amino acids. The shortest linkers are clearly related to one another and fall into two subgroups: (i) YkvD and KinB and (ii) YvfT, YocF, and YdfH (data not shown). The intermediate-length linkers from YclK, YvqE, YvqB, YrkQ, YesM, and YbdK are related and have a conserved sequence DEIGXhyA (hy is any hydrophobic residue) beginning about 40 residues distal to the last transmembrane region (Fig. 6). This sequence is also found in ResE and YycG. A second conserved sequence, GhyhyAhyhyXDXTE appears in the histidine proximal region of YufL, YbdF, CitS, YycG, ResE, PhoR, and KinC. Both of these conserved motifs may have something to do with the activity of the kinases, although that function remains obscure. In the case of KinC, a PAS domain is known to be present in the cytoplasmic linker, but neither motif would be included in the PAS domain (32). It seems likely that the motifs define a signal input site, perhaps to modulate the response to other signals. Their presence in a number of kinases suggests that the linker may be the target of a global regulatory system.
FIG. 6.
Similarities of sequences of cytoplasmic linkers. Sequences of linkers between the last transmembrane domain and the histidine motif of kinases that show homology are compared. The shaded residues define two motifs common to these linkers. Numerous partial homologies are not shaded for clarity. Gaps introduced to maximize alignment are indicated by the dots.
While the transmembrane and periplasmic subdomains of the sensor domain may indeed be ligand-binding signal input domains in many kinases, this need not be the case in all kinases. The cytoplasmic linker domain or even the histidine phosphotransferase domain itself could be sites of kinase activation or inhibition. In fact, deletion experiments with the PhoR kinase of B. subtilis revealed that the sensor domain is unnecessary for phosphate-regulated activation of PhoR activity (3). In some kinases, the periplasmic and transmembrane regions may serve other functions such as aggregation with specific proteins (16) or spatiotemporal placement in the cell membrane (25).
MOLECULAR BASIS FOR KINASE-RESPONSE REGULATOR SPECIFICITY
The multitude of kinase-response regulator pairs found in B. subtilis (14), E. coli (19), and Synechocystis (20) along with the structural conservation of response regulators and, most likely, the histidine phosphotransferase domains of kinases raises the question of how the cell ensures specific signals activate the right genes. There must be exquisite specificity of interaction between the kinase and its response regulator partner in order to exclude other response regulators from stealing the kinase phosphoryl group and activating inappropriate genes. Protein-protein interactions normally occur over fairly large surfaces and are multifactorial; i.e., many weak interactions are involved. The surfaces required for such interactions in two-component systems have been studied in CheA-CheY (31) and PhoR-PhoB (8) of E. coli as well as KinA-Spo0F of B. subtilis (29). Alanine-scanning mutagenesis studies of Spo0F indicate that the residues most important for kinase interaction surround the active-site aspartates. These residues were also implicated in the PhoB studies, while CheY may have more than one surface of interaction with CheA (18). It is virtually certain that the residues around the active-site aspartates must make productive interaction with residues around the phosphohistidine in all of the kinases. Because within each kinase group there are sequences around the histidine that differ only by one or two residues (Fig. 2), it was unclear how individual specificities are maintained within the group. To address this question, a comparison of the residues around the active-site aspartates of response regulators thought to be involved in the kinase-response regulator interaction surface was undertaken. These residues are contained within the loops connecting the β-sheets and α-helices, and mutation of these residues in suppressor studies or alanine-scanning studies is known to lead to altered kinase specificity or to affect kinase interaction.
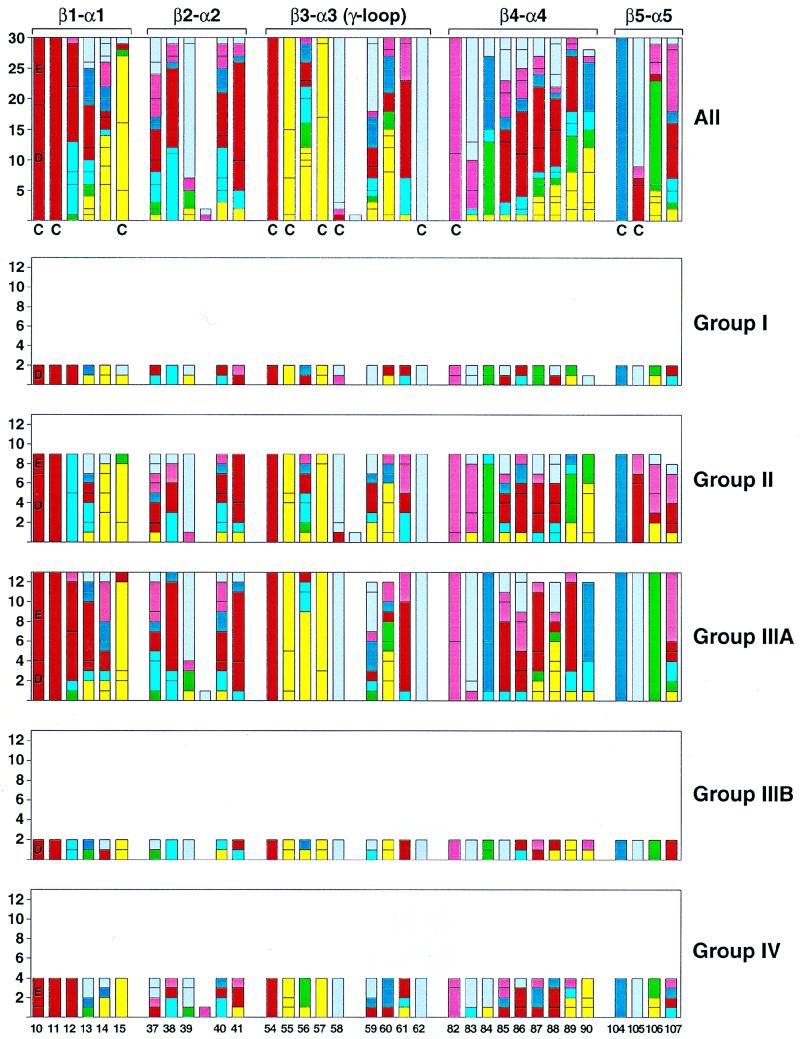
A compilation of the residues in the β-α loops within each family of response regulator is presented in Fig. 7. Although more detail is presented than can be interpreted here, some general conclusions may be drawn to help in this context. The β3-α3 (γ-loop) has the most conservative residues, and these are located distal from the phosphorylated aspartate (residue 54). Residues in this loop are important for Mg2+ coordination and for the stability of the active site. The two major groups of response regulators, groups II and IIIA, for which enough examples exist to make some generalizations, differ in some key residues. For example, the essential aspartate at position 11 is followed by a basic residue in group II and an acidic residue in group IIIA. The key lysine at position 104 is followed by a proline in all groups except group II where it mainly is an acidic residue. A major change such as this is likely to have consequences in the arrangement of the α1-α5 interface. Comparing groups II and IIIA, several other residues including residues 14, 83, 84, and 106 are conserved within a group and different from the other group. This suggests the concept that group or family specificities exist within response regulators that define a common interaction surface for the conserved structure of that group’s kinase histidine domains with which this surface must interact. Individual specificities within a family must arise from the nonconserved residues within the loops either singly or in combination with others. The major prediction from these conclusions is that single-amino-acid changes in response regulator residues involved in individual specificity are most likely to result in altered interaction with kinases of the same group. As a corollary, if cross talk between kinase-response regulator pairs is of regulatory significance, it is likely to occur only within a group.
FIG. 7.
Sequence alignment of response regulator loop regions proximal to the site of phosphorylation for B. subtilis. The number of each response regulator residue type at loop positions spanning between secondary structure elements β-strand 1 to α-helix 1 (β1-α1), β2-α2, β3-α3, β4-α4, and β5-α5 are illustrated individually as members of groups I, II, IIIA, and IIIB, IV and collectively in the top bar graph for comparison. The residue type is coded by color: acidic in red (D and E), basic in dark blue (K and R), hydroxyl in rose (S and T), polar in light blue (H, N, and Q), hydrophobic in yellow (C, I, L, V, and M), aromatic in green (Y, F, and W), and structural in gray (A, G, and P). The numbering scheme is based on the Spo0F sequence, and conserved residues essential for phosphorylation are D10, D11, D54 (the phosphorylation site), T82, and K104 by this numbering. Residues previously described as conserved (C) by alignment of response regulators from all organisms (30) are denoted. Response regulator sequences (14) were aligned with Clustal W (10) and illustrated with the Excel 98 (Microsoft) and Adobe Illustrator programs (Adobe).
PERSPECTIVES
It is now becoming clear how two-component systems work. The basic chemical mechanisms of phosphotransfer, the structure of active domains, and the requirements for histidine kinase-response regulator interaction are relatively easy to study, and therefore, much is known. With a few exceptions, the nature of the signals activating these kinases remains obscure. The structure of histidine kinases, with the exception of some kinase fragments, and the mechanism of signal-activated autophosphorylation remain mysteries. The cellular roles of most two-component systems and the genes they activate are unknown. It is safe to conclude that there is much to learn about bacterial responses to their environment and how these systems help to mediate that response.
ACKNOWLEDGMENT
This work was supported, in part, by grant GM19416 from the National Institute of General Medical Sciences, National Institutes of Health, USPHS.
Footnotes
†
Publication 12190-MEM from the Department of Molecular and Experimental Medicine at The Scripps Research Institute.
REFERENCES
- 1.Appleby J L, Parkinson J S, Bourret R B. Signal transduction via the multistep phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 2.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 3.Birkey S M, Liu W, Zhang X, Duggan M F, Hulett F M. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol Microbiol. 1998;30:943–953. doi: 10.1046/j.1365-2958.1998.01122.x. [DOI] [PubMed] [Google Scholar]
- 4.Burbulys D, Trach K A, Hoch J A. The initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 5.Dartois V, Débarbouillé M, Kunst F, Rapoport G. Characterization of a novel member of the DegS-DegU regulon affected by salt stress in Bacillus subtilis. J Bacteriol. 1998;180:1855–1861. doi: 10.1128/jb.180.7.1855-1861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egger L A, Park H, Inouye M. Signal transduction via the histidyl-aspartyl phosphorelay. Genes-Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 7.Fabret C, Hoch J A. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher S L, Jiang W, Wanner B L, Walsh C T. Cross-talk between the histidine protein kinase VanS and the response regulator PhoB. J Biol Chem. 1995;270:23143–23149. doi: 10.1074/jbc.270.39.23143. [DOI] [PubMed] [Google Scholar]
- 9.Grossman A D. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 10.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, M., M. Perego, and J. A. Hoch. Unpublished data.
- 12.Jiang, M., Y.-L. Tzeng, V. A. Feher, M. Perego, and J. A. Hoch. Unpublished data.
- 13.Kobayashi K, Shoji K, Shimizu T, Nakano K, Sato T, Kobayaski Y. Analysis of a suppressor mutation ssb (kinC) of sur0B20 (spo0A) mutation in Bacillus subtilis reveals that kinC encodes a histidine protein kinase. J Bacteriol. 1995;177:176–182. doi: 10.1128/jb.177.1.176-182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunst F, et al. The complete genome sequence of the Gram-positive model organism Bacillus subtilis (strain 168) Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 15.LeDeaux J R, Grossman A D. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177:166–175. doi: 10.1128/jb.177.1.166-175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levit M N, Liu Y, Stock J B. Stimulus response coupling in bacterial chemotaxis: receptor dimers in signaling arrays. Mol Microbiol. 1998;30:459–466. doi: 10.1046/j.1365-2958.1998.01066.x. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Hackert E, Stock A M. The DNA-binding domain of OmpR: crystal structure of a winged helix transcription factor. Structure. 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 18.McEvoy M M, Hausrath A C, Randolph G B, Remington S J, Dahlquist F W. Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway. Proc Natl Acad Sci USA. 1998;95:7333–7338. doi: 10.1073/pnas.95.13.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizuno T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997;4:161–168. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- 20.Mizuno T, Kaneko T, Tabata S. Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium, Synechocystis sp. strain PCC 6803. DNA Res. 1996;3:407–414. doi: 10.1093/dnares/3.6.407. [DOI] [PubMed] [Google Scholar]
- 21.Nakano M M, Zuber P, Glaser P, Danchin A, Hulett F M. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fns upon oxygen limitation in Bacillus subtilis. J Bacteriol. 1996;178:3796–3802. doi: 10.1128/jb.178.13.3796-3802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohlsen K L, Grimsley J K, Hoch J A. Deactivation of the sporulation transcription factor Spo0A by the Spo0E protein phosphatase. Proc Natl Acad Sci USA. 1994;91:1756–1760. doi: 10.1073/pnas.91.5.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 24.Rosario M M L, Ordal G W. CheC and CheD interact to regulate methylation of Bacillus subtilis methyl-accepting chemotaxis proteins. Mol Microbiol. 1996;21:511–518. doi: 10.1111/j.1365-2958.1996.tb02560.x. [DOI] [PubMed] [Google Scholar]
- 25.Shapiro L, Losick R. Protein localization and cell fate in bacteria. Science. 1997;276:712–718. doi: 10.1126/science.276.5313.712. [DOI] [PubMed] [Google Scholar]
- 26.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive response in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun G, Birkey S M, Hulett F M. Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol Microbiol. 1996;19:941–948. doi: 10.1046/j.1365-2958.1996.422952.x. [DOI] [PubMed] [Google Scholar]
- 28.Trach K A, Hoch J A. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol Microbiol. 1993;8:69–79. doi: 10.1111/j.1365-2958.1993.tb01204.x. [DOI] [PubMed] [Google Scholar]
- 29.Tzeng Y-L, Hoch J A. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J Mol Biol. 1997;272:200–212. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- 30.Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 31.Zhu X, Volz K, Matsumura P. The CheZ-binding surface of CheY overlaps the CheA- and FliM-binding surfaces. J Biol Chem. 1997;272:23758–23764. doi: 10.1074/jbc.272.38.23758. [DOI] [PubMed] [Google Scholar]
- 32.Zhulin I B, Taylor B L, Dixon R. PAS domain S-boxes in archaea, bacteria and sensors for oxygen and redox. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]