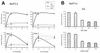Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5 - PubMed (original) (raw)
Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5
Anne Rascle et al. Mol Cell Biol. 2003 Jun.
Abstract
The signal transducer and activator of transcription STAT5 plays a major role in the cellular response to cytokines, but the mechanism by which it activates transcription remains poorly understood. We show here that deacetylase inhibitors (trichostatin A, suberoylanilide hydroxamic acid, and sodium butyrate) prevent induction of endogenous STAT5 target genes, implying that a deacetylase activity is required for that process. Microarray analyses revealed that this requirement is common to all STAT5 target genes. Using chromatin immunoprecipitation, we show that, following STAT5 DNA binding, deacetylase inhibitors block transcription initiation by preventing recruitment of the basal transcription machinery. This inhibition is not due to effects on histone H3 and H4 acetylation or chromatin remodeling within the promoter region. This novel mechanism of transactivation by STAT5 provides a rationale for the use of deacetylase inhibitors for therapeutic intervention in STAT5-associated cancers.
Figures
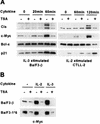
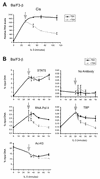
FIG. 1.
Cytokine induction of Cis and c-Myc proteins is abolished in TSA-treated murine lymphocytes. (A) Effects of TSA on protein expression in cytokine-stimulated murine B and T lymphocytes. Ba/F3-β and CTLL-2 were stimulated with IL-3 and IL-2, respectively, for the indicated times, as described in Materials and Methods. Cells were pretreated with 200 nM TSA (+) or DMSO (−) for 30 min prior to cytokine stimulation. Protein lysates were subjected to a Western blot analysis with the indicated antibodies. (B) c-Myc constitutive expression in a STAT5Aca cell line is repressed by TSA. Ba/F3-β and Ba/F3-1*6 were stimulated with IL-2 or IL-3 for 2 h in the presence of 200 nM TSA or DMSO, as described for panel A. Protein lysates were subjected to a Western blot analysis with a polyclonal antibody specific for c-Myc.
FIG. 2.
TSA inhibits induction of STAT5 target genes at the RNA level, through a primary effect. TSA specifically inhibits transcriptional induction of STAT5 target genes. (A and B) Ba/F3-β cells were stimulated with IL-3 in the absence or presence of TSA, as described in the legend to Fig. 1. mRNA levels were analyzed by real-time PCR, as described in Materials and Methods, with primers specific for STAT5 target genes (A) or control genes (B). (C) Inhibition of transcription by TSA does not require de novo protein synthesis. CTLL-2 cells were stimulated with IL-2 in the absence or presence of TSA and/or CHX, as described in Materials and Methods. mRNA levels were analyzed by real-time PCR as described above. (D) p21 expression is differentially affected by TSA in Ba/F3-β and CTLL-2 cells. Cells were treated, and mRNA levels were analyzed with primers specific for mouse p21, as described above.
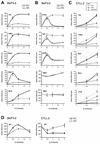
FIG. 3.
A deacetylase activity is required for cytokine induction of STAT5 target genes. (A) Three unrelated inhibitors of deacetylases block induction of STAT5 target genes. Ba/F3-β cells were stimulated with IL-3 in the presence of 200 nM TSA, 10 μM SAHA, 10 mM NaB, or DMSO (−), and mRNA levels were analyzed by real-time PCR, as described for Fig. 2. (B) Titration of TSA and SAHA. Ba/F3-β cells were stimulated with IL-3 for 30 min in the presence of increasing concentrations of TSA or SAHA, and mRNA levels were analyzed by real-time PCR, as described above. Data are expressed as the fold induction relative to untreated unstimulated cells.
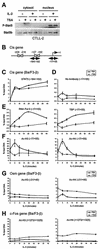
FIG. 4.
TSA blocks transcription initiation by preventing recruitment of the basal transcription machinery to the Cis promoter. (A) TSA does not interfere with STAT5 phosphorylation or its nuclear translocation upon cytokine stimulation. CTLL-2 cells were stimulated with IL-2 for 1 h in the absence or presence of TSA, as described for Fig. 2. Nuclear and cytosolic lysates were subjected to Western blot analysis with antibodies specific for phospho-STAT5 and STAT5b, as described in Materials and Methods. Identical results were obtained with a STAT5a-specific antibody (data not shown). (B) Structure of the murine Cis promoter. Gray boxes represent STAT5 binding sites, together with their positions relative to the transcription start site or CAP site (+1). Positions and sizes of the amplicons analyzed by ChIP in panels C to F are shown. (C to F) TSA does not interfere with STAT5 DNA binding but prevents histone acetylation and recruitment of TBP and RNA polymerase II at the CAP site of the Cis promoter. Ba/F3-β cells were stimulated with IL-3 in the absence or presence of TSA, as described for Fig. 1. At the times indicated, cells were harvested and analyzed by ChIP, with antibodies specific for STAT5a and -b (STAT5) (C), RNA polymerase II (RNA Pol II) and TBP (E), acetylated histone H3 (Ac-H3) and histone H4 (Ac-H4) (F), or no antibody as a control (D), as described in Materials and Methods. DNA samples were analyzed by real-time PCR, with primers amplifying the −184/−102 region (STAT5) or the −17/+55 CAP site region (RNA Pol II, TBP, Ac-H3, Ac-H4, and No Antibody). Identical results were obtained for STAT5 binding when the −256/−195 region was amplified (data not shown). (G and H) Effect of TSA on histone acetylation at the Osm and c-Fos genes. Ba/F3-β cells were treated, and histone acetylation was analyzed by ChIP, as described above, with primers specific for the CAP site (−21/+40, Osm) or the open reading frame (+1273/+1325, c-Fos). Histone acetylation in panels F to H is represented at identical scales for better comparison.
FIG. 5.
TSA inhibits transcription reinitiation by preventing reloading of the transcription machinery. (A) TSA rapidly inhibits Cis transcription in cytokine-stimulated cells. Ba/F3-β cells were stimulated with IL-3 in the absence of TSA for 30 min before either DMSO (−TSA) or 200 nM TSA (+TSA) was added to the cells (indicated by an arrowhead). Cells were harvested at the indicated times, and Cis mRNA levels were analyzed by real-time PCR. (B) TBP and RNA polymerase II are rapidly released from the Cis promoter upon TSA treatment of cytokine-stimulated cells. Ba/F3-β cells were treated as described for panel A, and cells were harvested at the indicated times and analyzed by ChIP, as described for Fig. 4C to F.
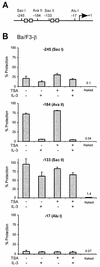
FIG. 6.
TSA does not affect chromatin remodeling on the Cis promoter. (A) Structure of the Cis promoter, indicating the positions of the restriction sites analyzed for panel B. Gray boxes represent STAT5 binding sites. (B) IL-3-mediated chromatin remodeling on the Cis promoter is not affected by TSA. Ba/F3-β cells were stimulated with IL-3 for 30 min in the absence or presence of 200 nM TSA, as described for Fig. 1. Unstimulated and stimulated cells were harvested, nuclei were prepared, and chromatin accessibility was analyzed by CHART-PCR, as described in Materials and Methods. Purified genomic (naked) DNA was included in a parallel reaction as a control for the cutting efficiency of the restriction enzyme. Data are expressed as the percentages of nondigested DNA (% Protection).
Similar articles
- Chromatin acetylation and remodeling at the Cis promoter during STAT5-induced transcription.
Rascle A, Lees E. Rascle A, et al. Nucleic Acids Res. 2003 Dec 1;31(23):6882-90. doi: 10.1093/nar/gkg907. Nucleic Acids Res. 2003. PMID: 14627821 Free PMC article. - NCoA-1/SRC-1 is an essential coactivator of STAT5 that binds to the FDL motif in the alpha-helical region of the STAT5 transactivation domain.
Litterst CM, Kliem S, Marilley D, Pfitzner E. Litterst CM, et al. J Biol Chem. 2003 Nov 14;278(46):45340-51. doi: 10.1074/jbc.M303644200. Epub 2003 Sep 3. J Biol Chem. 2003. PMID: 12954634 - Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription.
Nakajima H, Brindle PK, Handa M, Ihle JN. Nakajima H, et al. EMBO J. 2001 Dec 3;20(23):6836-44. doi: 10.1093/emboj/20.23.6836. EMBO J. 2001. PMID: 11726519 Free PMC article. - Signal transducer and activator of transcription 5 (STAT5).
Buitenhuis M, Coffer PJ, Koenderman L. Buitenhuis M, et al. Int J Biochem Cell Biol. 2004 Nov;36(11):2120-4. doi: 10.1016/j.biocel.2003.11.008. Int J Biochem Cell Biol. 2004. PMID: 15313458 Review. - Coactivators in gene regulation by STAT5.
Litterst CM, Kliem S, Lodrini M, Pfitzner E. Litterst CM, et al. Vitam Horm. 2005;70:359-86. doi: 10.1016/S0083-6729(05)70012-1. Vitam Horm. 2005. PMID: 15727811 Review.
Cited by
- Givinostat: an emerging treatment for polycythemia vera.
Chifotides HT, Bose P, Verstovsek S. Chifotides HT, et al. Expert Opin Investig Drugs. 2020 Jun;29(6):525-536. doi: 10.1080/13543784.2020.1761323. Epub 2020 Jul 21. Expert Opin Investig Drugs. 2020. PMID: 32693648 Free PMC article. Review. - A Pan-Histone Deacetylase Inhibitor Enhances the Antitumor Activity of B7-H3-Specific CAR T Cells in Solid Tumors.
Lei X, Ou Z, Yang Z, Zhong J, Zhu Y, Tian J, Wu J, Deng H, Lin X, Peng Y, Li B, He L, Tu Z, Chen W, Li Q, Liu N, Zhang H, Wang Z, Fang Z, Yamada T, Lv X, Tian T, Pan G, Wu F, Xiao L, Zhang L, Cai T, Wang X, Tannous BA, Li J, Kontos F, Ferrone S, Fan S. Lei X, et al. Clin Cancer Res. 2021 Jul 1;27(13):3757-3771. doi: 10.1158/1078-0432.CCR-20-2487. Epub 2021 Apr 2. Clin Cancer Res. 2021. PMID: 33811153 Free PMC article. - The synthetic α-bromo-2',3,4,4'-tetramethoxychalcone (α-Br-TMC) inhibits the JAK/STAT signaling pathway.
Pinz S, Unser S, Brueggemann S, Besl E, Al-Rifai N, Petkes H, Amslinger S, Rascle A. Pinz S, et al. PLoS One. 2014 Mar 3;9(3):e90275. doi: 10.1371/journal.pone.0090275. eCollection 2014. PLoS One. 2014. PMID: 24595334 Free PMC article. - HDAC6 Deacetylates HMGN2 to Regulate Stat5a Activity and Breast Cancer Growth.
Medler TR, Craig JM, Fiorillo AA, Feeney YB, Harrell JC, Clevenger CV. Medler TR, et al. Mol Cancer Res. 2016 Oct;14(10):994-1008. doi: 10.1158/1541-7786.MCR-16-0109. Epub 2016 Jun 29. Mol Cancer Res. 2016. PMID: 27358110 Free PMC article. - Histone deacetylase inhibitors suppress the expression of inflammatory and innate immune response genes in human microglia and astrocytes.
Suh HS, Choi S, Khattar P, Choi N, Lee SC. Suh HS, et al. J Neuroimmune Pharmacol. 2010 Dec;5(4):521-32. doi: 10.1007/s11481-010-9192-0. Epub 2010 Feb 17. J Neuroimmune Pharmacol. 2010. PMID: 20157787 Free PMC article.
References
- Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. - PubMed
- Ahringer, J. 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16:351-356. - PubMed
- Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. - PubMed
- Brondello, J. M., A. Brunet, J. Pouyssegur, and F. R. McKenzie. 1997. The dual specificity mitogen-activated protein kinase phosphatase-1 and -2 are induced by the p42/p44MAPK cascade. J. Biol. Chem. 272:1368-1376. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous