Fast and sensitive multiple alignment of large genomic sequences - PubMed (original) (raw)
Comparative Study
Fast and sensitive multiple alignment of large genomic sequences
Michael Brudno et al. BMC Bioinformatics. 2003.
Abstract
Background: Genomic sequence alignment is a powerful method for genome analysis and annotation, as alignments are routinely used to identify functional sites such as genes or regulatory elements. With a growing number of partially or completely sequenced genomes, multiple alignment is playing an increasingly important role in these studies. In recent years, various tools for pair-wise and multiple genomic alignment have been proposed. Some of them are extremely fast, but often efficiency is achieved at the expense of sensitivity. One way of combining speed and sensitivity is to use an anchored-alignment approach. In a first step, a fast search program identifies a chain of strong local sequence similarities. In a second step, regions between these anchor points are aligned using a slower but more accurate method.
Results: Herein, we present CHAOS, a novel algorithm for rapid identification of chains of local pair-wise sequence similarities. Local alignments calculated by CHAOS are used as anchor points to improve the running time of DIALIGN, a slow but sensitive multiple-alignment tool. We show that this way, the running time of DIALIGN can be reduced by more than 95% for BAC-sized and longer sequences, without affecting the quality of the resulting alignments. We apply our approach to a set of five genomic sequences around the stem-cell-leukemia (SCL) gene and demonstrate that exons and small regulatory elements can be identified by our multiple-alignment procedure.
Conclusion: We conclude that the novel CHAOS local alignment tool is an effective way to significantly speed up global alignment tools such as DIALIGN without reducing the alignment quality. We likewise demonstrate that the DIALIGN/CHAOS combination is able to accurately align short regulatory sequences in distant orthologues.
Figures
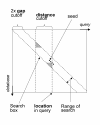
Figure 1
The figure shows a matrix representation of sequence alignment. The seed shown can be chained to any seed which lies inside the search box. All seeds located less then distance bp from the current location are stored in a skip list, in which we do a range query for seeds located within a gap cutoff from the diagonal on which the current seed is located. The seeds located in the grey areas are not available for chaining to make the algorithm independent of sequence order.
Figure 2
CHAOS-DIALIGN correctly aligns the SCL promoter and a conserved non-coding sequence in exon 1. The alignment was extracted from the CHAOS-DIALIGN global alignment of SCL sequences from human, mouse, chicken, zebrafish, and pufferfish. Consensus binding motifs are labelled. All except YY1 have been previously demonstrated to be essential for the appropriate pattern or level of SCL expression. The factors binding conserved sequence (CS) 1 and 2 are unknown. Shading of bases is at (grey) and (black) conservation.
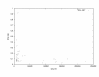
Figure 3
Relative improvement in program running time for 42 pairs of genomic sequences form human and mouse of different length. Each point represents one sequence pair. The _x_-axis is the medium sequence length of sequence pairs while the _y_-axis is the relative running time of the anchored-alignment procedure compared to the non-anchored procedure.
Similar articles
- Fast and sensitive alignment of large genomic sequences.
Brudno M, Morgenstern B. Brudno M, et al. Proc IEEE Comput Soc Bioinform Conf. 2002;1:138-47. Proc IEEE Comput Soc Bioinform Conf. 2002. PMID: 15838131 - The CHAOS/DIALIGN WWW server for multiple alignment of genomic sequences.
Brudno M, Steinkamp R, Morgenstern B. Brudno M, et al. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W41-4. doi: 10.1093/nar/gkh361. Nucleic Acids Res. 2004. PMID: 15215346 Free PMC article. - DIALIGN P: fast pair-wise and multiple sequence alignment using parallel processors.
Schmollinger M, Nieselt K, Kaufmann M, Morgenstern B. Schmollinger M, et al. BMC Bioinformatics. 2004 Sep 9;5:128. doi: 10.1186/1471-2105-5-128. BMC Bioinformatics. 2004. PMID: 15357879 Free PMC article. - Computation and analysis of genomic multi-sequence alignments.
Blanchette M. Blanchette M. Annu Rev Genomics Hum Genet. 2007;8:193-213. doi: 10.1146/annurev.genom.8.080706.092300. Annu Rev Genomics Hum Genet. 2007. PMID: 17489682 Review. - Sequence alignment in molecular biology.
Apostolico A, Giancarlo R. Apostolico A, et al. J Comput Biol. 1998 Summer;5(2):173-96. doi: 10.1089/cmb.1998.5.173. J Comput Biol. 1998. PMID: 9672827 Review.
Cited by
- The complex set of internal repeats in SpTransformer protein sequences result in multiple but limited alternative alignments.
Barela Hudgell MA, Smith LC. Barela Hudgell MA, et al. Front Immunol. 2022 Oct 18;13:1000177. doi: 10.3389/fimmu.2022.1000177. eCollection 2022. Front Immunol. 2022. PMID: 36330505 Free PMC article. - Multiple genome alignment in the telomere-to-telomere assembly era.
Kille B, Balaji A, Sedlazeck FJ, Nute M, Treangen TJ. Kille B, et al. Genome Biol. 2022 Aug 29;23(1):182. doi: 10.1186/s13059-022-02735-6. Genome Biol. 2022. PMID: 36038949 Free PMC article. Review. - Multiple Alignment of Promoter Sequences from the Arabidopsis thaliana L. Genome.
Korotkov EV, Suvorova YM, Kostenko DO, Korotkova MA. Korotkov EV, et al. Genes (Basel). 2021 Jan 21;12(2):135. doi: 10.3390/genes12020135. Genes (Basel). 2021. PMID: 33494278 Free PMC article. - A fast adaptive algorithm for computing whole-genome homology maps.
Jain C, Koren S, Dilthey A, Phillippy AM, Aluru S. Jain C, et al. Bioinformatics. 2018 Sep 1;34(17):i748-i756. doi: 10.1093/bioinformatics/bty597. Bioinformatics. 2018. PMID: 30423094 Free PMC article. - Accurate multiple alignment of distantly related genome sequences using filtered spaced word matches as anchor points.
Leimeister CA, Dencker T, Morgenstern B. Leimeister CA, et al. Bioinformatics. 2019 Jan 15;35(2):211-218. doi: 10.1093/bioinformatics/bty592. Bioinformatics. 2019. PMID: 29992260 Free PMC article.
References
- Chain P, Kurtz S, Ohlebusch E, Slezak T. An applications-focused review of comparative genomics tools: capabilities, limitations, and future challenges. Briefings in Bioinformatics. 2003;4:105–123. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous