Ectodomain shedding of the glycoprotein GP of Ebola virus - PubMed (original) (raw)
Ectodomain shedding of the glycoprotein GP of Ebola virus
Olga Dolnik et al. EMBO J. 2004.
Abstract
In this study, release of abundant amounts of the Ebola virus (EBOV) surface glycoprotein GP in a soluble form from virus-infected cells was investigated. We demonstrate that the mechanism responsible for the release of GP is ectodomain shedding mediated by cellular sheddases. Proteolytic cleavage taking place at amino-acid position D637 removes the transmembrane anchor and liberates complexes consisting of GP1 and truncated GP2 (GP(2delta)) subunits from the cell surface. We show that tumor necrosis factor alpha-converting enzyme (TACE), a member of the ADAM family of zinc-dependent metalloproteases, is involved in EBOV GP shedding. This finding shows for the first time that virus-encoded surface glycoproteins are substrates for ADAMs. Furthermore, we provide evidence that shed GP is present in significant amounts in the blood of virus-infected animals and that it may play an important role in the pathogenesis of infection by efficiently blocking the activity of virus-neutralizing antibodies.
Figures
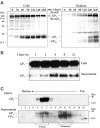
Figure 1
Release of soluble EBOV GP1,2Δ complexes. (A) RK-13 cells were infected with vSCGP8, pulse labeled 10 h p.i. and chased for 6 h. Culture medium (m) and cells (c) were collected separately. Soluble (s) and membrane-bound (p) proteins were separated by ultracentrifugation (25 000 rpm, 1 h, 4°C). Labeled proteins were immunoprecipitated using rabbit anti-GP2 Igs or human monoclonal antibody KZ52. (B) Western blot analysis of culture medium from EBOV-infected Vero E6, HeLa, and RK-13 cells.
Figure 2
Analysis of glycosylation pattern of GP2 and GP2Δ. (A) RK-13 cells were infected with vSCGP8 (wtGP) or vaccinia viruses expressing glycosylation mutants (GP618N/T and GP563N/T) and subjected to pulse-chase labeling and immunoprecipitation with horse anti-EBOV Igs followed by endoglycosidase treatment with Endo H or PNGase F. Forms of GP2 differing in N-glycosylation pattern are indicated as follows: A—containing only complex type oligosaccharides, B—containing only high-mannose type oligosaccharides, C—completely deglycosylated molecules. (B) Culture medium from Vero E6 cells infected with EBOV was ultracentrifuged, and pellet and supernatant were collected separately. Samples were either treated with Endo H or PNGase F or used untreated. Proteins were analyzed by Western blot.
Figure 3
Kinetics of release and oligomeric structure of soluble GP. (A) RK-13 cells infected with vSCGP8 were pulse-chase labeled and proteins were immunoprecipitated from cells (left panel) and medium (right panel). (B) Surface biotinylation of RK-13 cells expressing EBOV GP. Release of biotin-labeled GP from the cell surface into culture supernatant was observed at indicated time intervals. (C) Analysis of the oligomeric structure of shed GP. Vero E6 cells were infected with EBOV. At 5 days p.i., virus from culture medium was pelleted through a 20% sucrose cushion, and subsequently the supernatant and virus were analyzed by ultracentrifugation at linear sucrose gradients in the presence of 1% NP-40 and 0.4% DOC. Fractions 1–15 were collected from the bottom and analyzed by Western blot. Due to the effect of several detergents present in the samples such as NP-40, DOC, and finally SDS, the bands appeared slightly more diffused compared to other analyses.
Figure 4
Determination of the carboxy-terminus of GP2Δ. (A) Schematic illustration of EBOV GP2. Subtilisin-like cleavage site (RTRR), shedding cleavage site (D637–Q638), position of carboxy-terminal deletion mutants (ΔTM, Δ630, Δ640), cysteine residues (C), N-glycosylation sites N563 and N618 (white stars), transmembrane anchor (TM), cytoplasmic tail (CT), and fusion peptide (FD) are indicated. (B) Western blot analysis of GP deletion mutants. Culture medium from HeLa cells infected with vTF7-3 and transfected with pGEMmGP8/ΔTM, pGEMmGP8/Δ630, or pGEMmGP8/Δ640 was treated with PNGase F and subjected to Western blot analysis. GP2 from virus particles (p) and shed GP2Δ from EBOV-infected HeLa cells (s) were used as controls. (C) SDS–PAGE analysis of purified soluble GP under nonreducing conditions. The left panel shows silver staining and the right panel immunodetection with anti-GP2 Igs. (D) Analysis of purified GP preparation by MALDI-TOF MS. GP2Δ and PNGase F peaks are indicated. Upper box: GP2Δ mass after subtraction of carboxyamidoyl group mass (CAM); lower box: theoretical masses of GP2Δ with indicated carboxy-terminal amino acid. (E) Influence of amino-acid exchanges on GP shedding. RK-13 cells infected with vSCGP8 (wtGP) or virus mutants with indicated exchanges were labeled and analyzed by immunoprecipitation. Quantification of released GP2Δ was performed using Bio-imaging. Representative results from individual experiments are presented as mean values (±s.d.).
Figure 5
Involvement of TACE in EBOV GP shedding. (A) 293 cells were transfected with pcDNA3.1-GP or co-transfected with pcDNA3.1-GP and pcDNA3Mu TACE FL. At 36 h p.t., cells were pulse labeled and chased for 8 h. Labeled proteins were analyzed by immunoprecipitation. (B) 293 cells were infected with vSCGP8 and subsequently transfected with ASOs. Mismatch ASO2M and ASO4M were used as negative controls. At 7 h p.t., cells were labeled and chased for 8 h. Proteins from culture medium were analyzed by immunoprecipitation. (C) EC-2 cells (TACEΔZn/ΔZn) lacking functional TACE were transfected with pcDNA3.1-GP or co-transfected with pcDNA3Mu TACE FL and pcDNA3.1-GP. Cells were pulse-chase labeled 18 h p.t., and medium and cells were analyzed by immunoprecipitation. For all three panels (A–C) statistical analysis using at least three independent experiments has been performed. Mean values of GP release (% ±s.d.) are shown in bars. Quantification of the released GP2Δ was performed using Fuji BAS 1000 Bio-Imaging Analyzer.
Figure 6
GP1,2Δ is present in the blood of infected guinea pigs and is able to inhibit the virus-neutralizing activity of human monoclonal antibody KZ52. (A) Virus neutralization inhibition assays were performed using different EBOV dilutions in the presence of KZ52 antibody or shed GP or both at indicated concentrations. Total number of infectious units obtained for each reaction is indicated in bars. (B) Vero E6 cells were infected with EBOV and culture medium collected 5 days p.i. (EBOV-m). Guinea pigs (strain Hartley) were infected intraperitoneally with 1 × 103 PFU/animal of guinea pig-adapted EBOV (Volchkov et al, 2000). At days 6 and 9 p.i. (EBOV-6d, EBOV-9d), GP2, GP2Δ, and VP40 were identified in sera (sr) and in culture medium (m) by Western blot analysis using specific antibodies. Samples from blood (at day 9 p.i.) and culture medium were either treated with Endo H or PNGase F or used untreated.
Similar articles
- Uncoupling GP1 and GP2 expression in the Lassa virus glycoprotein complex: implications for GP1 ectodomain shedding.
Illick MM, Branco LM, Fair JN, Illick KA, Matschiner A, Schoepp R, Garry RF, Guttieri MC. Illick MM, et al. Virol J. 2008 Dec 23;5:161. doi: 10.1186/1743-422X-5-161. Virol J. 2008. PMID: 19105844 Free PMC article. - MARCH8 Inhibits Ebola Virus Glycoprotein, Human Immunodeficiency Virus Type 1 Envelope Glycoprotein, and Avian Influenza Virus H5N1 Hemagglutinin Maturation.
Yu C, Li S, Zhang X, Khan I, Ahmad I, Zhou Y, Li S, Shi J, Wang Y, Zheng YH. Yu C, et al. mBio. 2020 Sep 15;11(5):e01882-20. doi: 10.1128/mBio.01882-20. mBio. 2020. PMID: 32934085 Free PMC article. - The Tetherin Antagonism of the Ebola Virus Glycoprotein Requires an Intact Receptor-Binding Domain and Can Be Blocked by GP1-Specific Antibodies.
Brinkmann C, Nehlmeier I, Walendy-Gnirß K, Nehls J, González Hernández M, Hoffmann M, Qiu X, Takada A, Schindler M, Pöhlmann S. Brinkmann C, et al. J Virol. 2016 Nov 28;90(24):11075-11086. doi: 10.1128/JVI.01563-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27707924 Free PMC article. - The Roles of Ebola Virus Soluble Glycoprotein in Replication, Pathogenesis, and Countermeasure Development.
Zhu W, Banadyga L, Emeterio K, Wong G, Qiu X. Zhu W, et al. Viruses. 2019 Oct 31;11(11):999. doi: 10.3390/v11110999. Viruses. 2019. PMID: 31683550 Free PMC article. Review. - Shedding of membrane proteins by ADAM family proteases.
Moss ML, Lambert MH. Moss ML, et al. Essays Biochem. 2002;38:141-53. doi: 10.1042/bse0380141. Essays Biochem. 2002. PMID: 12463167 Review.
Cited by
- Forty-five years of Marburg virus research.
Brauburger K, Hume AJ, Mühlberger E, Olejnik J. Brauburger K, et al. Viruses. 2012 Oct 1;4(10):1878-927. doi: 10.3390/v4101878. Viruses. 2012. PMID: 23202446 Free PMC article. Review. - The Role of Conserved N-Linked Glycans on Ebola Virus Glycoprotein 2.
Lennemann NJ, Walkner M, Berkebile AR, Patel N, Maury W. Lennemann NJ, et al. J Infect Dis. 2015 Oct 1;212 Suppl 2(Suppl 2):S204-9. doi: 10.1093/infdis/jiv201. Epub 2015 Jun 2. J Infect Dis. 2015. PMID: 26038399 Free PMC article. - Molecular and cellular mechanisms of ectodomain shedding.
Hayashida K, Bartlett AH, Chen Y, Park PW. Hayashida K, et al. Anat Rec (Hoboken). 2010 Jun;293(6):925-37. doi: 10.1002/ar.20757. Anat Rec (Hoboken). 2010. PMID: 20503387 Free PMC article. Review. - Steric shielding of surface epitopes and impaired immune recognition induced by the ebola virus glycoprotein.
Francica JR, Varela-Rohena A, Medvec A, Plesa G, Riley JL, Bates P. Francica JR, et al. PLoS Pathog. 2010 Sep 9;6(9):e1001098. doi: 10.1371/journal.ppat.1001098. PLoS Pathog. 2010. PMID: 20844579 Free PMC article. - Uncoupling GP1 and GP2 expression in the Lassa virus glycoprotein complex: implications for GP1 ectodomain shedding.
Illick MM, Branco LM, Fair JN, Illick KA, Matschiner A, Schoepp R, Garry RF, Guttieri MC. Illick MM, et al. Virol J. 2008 Dec 23;5:161. doi: 10.1186/1743-422X-5-161. Virol J. 2008. PMID: 19105844 Free PMC article.
References
- Althoff K, Reddy P, Voltz N, Rose-John S, Müllberg J (2000) Shedding of interleukin 6 receptor and tumor necrosis factor alpha: contribution of the stalk sequence to the cleavage pattern of transmembrane proteins. Eur J Biochem 267: 2624–2631 - PubMed
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP (1997) A metalloproteinase desintegrin that releases tumor-necrosis factor-alpha from cells. Nature 385: 729–733 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous