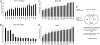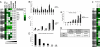Chromosomal instability determines taxane response - PubMed (original) (raw)
. 2009 May 26;106(21):8671-6.
doi: 10.1073/pnas.0811835106. Epub 2009 May 19.
Barbara Nicke, Marion Schuett, Aron C Eklund, Charlotte Ng, Qiyuan Li, Thomas Hardcastle, Alvin Lee, Rajat Roy, Philip East, Maik Kschischo, David Endesfelder, Paul Wylie, Se Nyun Kim, Jie-Guang Chen, Michael Howell, Thomas Ried, Jens K Habermann, Gert Auer, James D Brenton, Zoltan Szallasi, Julian Downward
Affiliations
- PMID: 19458043
- PMCID: PMC2688979
- DOI: 10.1073/pnas.0811835106
Chromosomal instability determines taxane response
Charles Swanton et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Microtubule-stabilizing (MTS) agents, such as taxanes, are important chemotherapeutics with a poorly understood mechanism of action. We identified a set of genes repressed in multiple cell lines in response to MTS agents and observed that these genes are overexpressed in tumors exhibiting chromosomal instability (CIN). Silencing 22/50 of these genes, many of which are involved in DNA repair, caused cancer cell death, suggesting that these genes are involved in the survival of aneuploid cells. Overexpression of these "CIN-survival" genes is associated with poor outcome in estrogen receptor-positive breast cancer and occurs frequently in basal-like and Her2-positive cases. In diploid cells, but not in chromosomally unstable cells, paclitaxel causes repression of CIN-survival genes, followed by cell death. In the OV01 ovarian cancer clinical trial, a high level of CIN was associated with taxane resistance but carboplatin sensitivity, indicating that CIN may determine MTS response in vivo. Thus, pretherapeutic assessment of CIN may optimize treatment stratification and clinical trial design using these agents.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
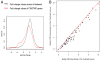
Fig. 1.
Genes overexpressed in tumors with CIN are repressed by MTS treatment in vitro and in vivo. (A) The CIN27wp gene expression signature, which correlates with total functional aneuploidy in several cancer types (9), was analyzed in 5 gene expression data sets measuring response to MTS treatment. The distribution of changes induced by MTS was lower in the CIN27wp genes (red) compared with the set of all measured genes (black) (P = 3.3e-7). (B) As part of the OV01 clinical trial, expression profiling was performed on ovarian carcinomas before and after 3 cycles of paclitaxel (Px) treatment. For each gene in the CIN70 signature, the median expression in the pretreatment and posttreatment tumors was compared, with the red line indicating equivalence.
Fig. 2.
Identification of 22 CIN-survival genes. (A) siRNA silencing of MTS-repressed genes impairs cell viability. Genes repressed within the 2 MTS expression signatures that are overexpressed in CIN tumors significantly altered HCT-116 cell viability when targeted by siRNA. An Acumen eX3 cytometer (Acumen TTP Labtech) was used to quantify viable cells 72 h after siRNA transfection. SDs are displayed for 3 independent experiments. P values are shown for Student 2-sided _t_-tests in all cases: *P < 0.05; **P < 0.005; ***P < 0.0005. (B) siRNA silencing of MTS-repressed genes promotes cell death. FACS analysis was used to quantify the mean subG1 fraction 72 h after transfection of siRNA. SDs and P values are displayed for 3 independent experiments. (C and D) Identification of 22 CIN-survival genes. Cells were transfected with siRNA targeting the 50 genes overexpressed in CIN tumors and repressed by MTS agents. Cell viability was quantified relative to scrambled control siRNA by a CellTiter-Blue assay 4 days after siRNA transfection in triplicate. Shown are the 22 genes that significantly impaired viability in the MDA-MB-468 and A549 cell lines. (E) Flowchart of analysis resulting in the derivation of 22 CIN-survival genes. The binomial test with probability corrections (BTPC) and rank methods were used to derive 2 MTS expression signatures. A total of 50 genes were repressed in the MTS signatures and overexpressed in CIN tumors. Of these 50 genes, 22 impaired cancer cell viability and/or induced apoptosis when silenced by RNA interference in 3 cancer cell lines: HCT-116 (colon), A549 (NSCLC), and MDA-MB-468 (breast).
Fig. 3.
Paclitaxel cytotoxicity is associated with CIN-survival gene repression. (A) Quantification of gene repression following paclitaxel treatment of cell lines with increasing chromosomal numerical heterogeneity. Fold change in gene expression post-paclitaxel treatment (24 h) relative to expression in vehicle control treated cells was determined by qPCR analysis (normalization to 18S and GAPDH) after 24 h of paclitaxel treatment (50% of the Gi50 concentration) in 3 biological replicate experiments in cell lines with increasing CIN. Color-coding represents the mean fold change in gene expression of 3 biological replicates relative to cells treated with vehicle alone + 1 SD. CIN-survival genes are highlighted in gray. Gene expression following paclitaxel treatment is compared with the HCT-116 cell line displaying the lowest CIN. The significance of the differences in gene expression were determined using the unpaired 2-sided Student _t_-test: *P <0.05; **P <0.005. (B) CIN-survival genes are relatively overexpressed in CINhigh COLO205 and SW620 cells compared with near-diploid CINlow HCT-116 cells. Shown is the relative quantification of gene expression normalized to 18S and GAPDH in COLO205 and SW620 cells compared with near-diploid HCT-116 cells. The graph indicates the mean fold change in gene expression compared with HCT-116 cells of 3 biological replicates (+ 1 SD). (C) Colorectal cancer cell lines with increasing chromosomal numerical heterogeneity uncouple mitotic arrest from cell death. Cell lines were treated with paclitaxel relative to the Gi50, Gi50/2, and Gi50/4 concentrations. Quantification of dying cells (subG1 fraction) and cells arrested in mitosis (MPM2-positive fraction) were assessed by FACS analysis. The MPM2:subG1 ratio was calculated by assessing the percentage of MPM2 positive cells relative to the percentage of subG1 cells following 24 h of paclitaxel treatment. The mean ratio of 3 independent experiments is presented (+1 SD). Cell lines are represented in ascending order of CIN status (21). (D and E) Silencing CIN-survival genes in the SW620 CINhigh cell line promotes cell death after 24 h of paclitaxel Gi50 treatment. At 48 h after transfection of CIN-survival siRNAs, SW620 cells were treated with paclitaxel for 24 h, and cells were prepared and analyzed as in C. In D, the mean MPM2:subG1 ratio of 2 independent experiments is presented (+ 1 SD). E shows that silencing of NUP205, H2AFX, CDC6, and RPA1 promoted a significant increase in subG1 cells after paclitaxel exposure compared with DMSO-treated cells (P <0.05; Student _t_-test). (F) Impaired repression of CIN-survival genes following paclitaxel treatment in an isogenic model of CIN. Quantification of gene repression by qPCR analysis of 21 CIN-survival genes following treatment of HCT-116 wild-type parental cells and isogenic Mad2+/- cells after 24 h of paclitaxel treatment (50 nM) normalized to 18s. Color-coding represents the mean fold repression + 1 SD from 3 biological replicates. P values indicate significantly greater gene repression in the HCT-116 parental cell line (Student 1-sided _t_-test). (G) CIN-survival gene repression correlates with cytotoxic response and stable tumor karyotype.
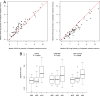
Fig. 4.
CIN70 predicts paclitaxel sensitivity and is a surrogate for CIN in breast cancer. (A) Expression of CIN70 genes determines sensitivity to paclitaxel and carboplatin. The figure contrasts basal median gene expression for each CIN70 gene in tumors with differing responses to paclitaxel and carboplatin. Paclitaxel-resistant tumors exhibited a higher median log-intensity of the CIN70 signature compared with paclitaxel-sensitive tumors (P = 0.043). CIN70 gene expression differed significantly between tumors subsequently resistant to paclitaxel and tumors resistant to carboplatin (P = 0.044; Student 2-sided _t_-test). (B) Expression of CIN and CIN-survival genes was greater in the aGU breast cancers. DNA image cytometry was used to classify breast cancers as dGS, aGS, or aGU. Boxplots summarize the expression of the CIN70, CIN27wp, and CIN-survival signatures within each group. P values were calculated using the Student 2-tailed _t_-test.
Similar articles
- Taxane benefit in breast cancer--a role for grade and chromosomal stability.
A'Hern RP, Jamal-Hanjani M, Szász AM, Johnston SR, Reis-Filho JS, Roylance R, Swanton C. A'Hern RP, et al. Nat Rev Clin Oncol. 2013 Jun;10(6):357-64. doi: 10.1038/nrclinonc.2013.67. Epub 2013 May 7. Nat Rev Clin Oncol. 2013. PMID: 23648828 Review. - A semisynthetic taxane Yg-3-46a effectively evades P-glycoprotein and β-III tubulin mediated tumor drug resistance in vitro.
Cai P, Lu P, Sharom FJ, Fang WS. Cai P, et al. Cancer Lett. 2013 Dec 1;341(2):214-23. doi: 10.1016/j.canlet.2013.08.010. Epub 2013 Aug 11. Cancer Lett. 2013. PMID: 23941826 - Chromosomal instability, colorectal cancer and taxane resistance.
Swanton C, Tomlinson I, Downward J. Swanton C, et al. Cell Cycle. 2006 Apr;5(8):818-23. doi: 10.4161/cc.5.8.2682. Epub 2006 Apr 17. Cell Cycle. 2006. PMID: 16628000 Review. - Blocking Mitotic Exit of Ovarian Cancer Cells by Pharmaceutical Inhibition of the Anaphase-Promoting Complex Reduces Chromosomal Instability.
Raab M, Sanhaji M, Zhou S, Rödel F, El-Balat A, Becker S, Strebhardt K. Raab M, et al. Neoplasia. 2019 Apr;21(4):363-375. doi: 10.1016/j.neo.2019.01.007. Epub 2019 Mar 7. Neoplasia. 2019. PMID: 30851646 Free PMC article. - Live Imaging to Study Microtubule Dynamic Instability in Taxane-resistant Breast Cancers.
Wang R, Wang H, Wang Z. Wang R, et al. J Vis Exp. 2017 Feb 20;(120):55027. doi: 10.3791/55027. J Vis Exp. 2017. PMID: 28287508 Free PMC article.
Cited by
- Taxane benefit in breast cancer--a role for grade and chromosomal stability.
A'Hern RP, Jamal-Hanjani M, Szász AM, Johnston SR, Reis-Filho JS, Roylance R, Swanton C. A'Hern RP, et al. Nat Rev Clin Oncol. 2013 Jun;10(6):357-64. doi: 10.1038/nrclinonc.2013.67. Epub 2013 May 7. Nat Rev Clin Oncol. 2013. PMID: 23648828 Review. - Novel actions of next-generation taxanes benefit advanced stages of prostate cancer.
de Leeuw R, Berman-Booty LD, Schiewer MJ, Ciment SJ, Den RB, Dicker AP, Kelly WK, Trabulsi EJ, Lallas CD, Gomella LG, Knudsen KE. de Leeuw R, et al. Clin Cancer Res. 2015 Feb 15;21(4):795-807. doi: 10.1158/1078-0432.CCR-14-1358. Clin Cancer Res. 2015. PMID: 25691773 Free PMC article. - Losing balance: the origin and impact of aneuploidy in cancer.
Holland AJ, Cleveland DW. Holland AJ, et al. EMBO Rep. 2012 Jun 1;13(6):501-14. doi: 10.1038/embor.2012.55. EMBO Rep. 2012. PMID: 22565320 Free PMC article. Review. - The progress in our understanding of CIN in breast cancer research.
Liao YY, Cao WM. Liao YY, et al. Front Oncol. 2023 Feb 16;13:1067735. doi: 10.3389/fonc.2023.1067735. eCollection 2023. Front Oncol. 2023. PMID: 36874134 Free PMC article. Review. - Role of prolonged mitotic checkpoint activation in the formation and treatment of cancer.
Dalton WB, Yang VW. Dalton WB, et al. Future Oncol. 2009 Nov;5(9):1363-70. doi: 10.2217/fon.09.118. Future Oncol. 2009. PMID: 19903065 Free PMC article. Review.
References
- Weaver BA, Cleveland DW. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8:7–12. - PubMed
- Tao W, et al. Induction of apoptosis by an inhibitor of the mitotic kinesin KSP requires both activation of the spindle assembly checkpoint and mitotic slippage. Cancer Cell. 2005;8:49–59. - PubMed
- Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. - PubMed
- Swanton C, Tomlinson I, Downward J. Chromosomal instability, colorectal cancer and taxane resistance. Cell Cycle. 2006;5:818–823. - PubMed
- Sudo T, Nitta M, Saya H, Ueno NT. Dependence of paclitaxel sensitivity on a functional spindle assembly checkpoint. Cancer Res. 2004;64:2502–2508. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0701935/MRC_/Medical Research Council/United Kingdom
- P50 CA089393/CA/NCI NIH HHS/United States
- R21LM008823-01A1/LM/NLM NIH HHS/United States
- R21 LM008823/LM/NLM NIH HHS/United States
- P50 CA 89393/CA/NCI NIH HHS/United States
- CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous