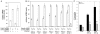Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells - PubMed (original) (raw)
Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells
Vincent M Bruno et al. PLoS Pathog. 2009 Aug.
Abstract
Recognition of conserved bacterial products by innate immune receptors leads to inflammatory responses that control pathogen spread but that can also result in pathology. Intestinal epithelial cells are exposed to bacterial products and therefore must prevent signaling through innate immune receptors to avoid pathology. However, enteric pathogens are able to stimulate intestinal inflammation. We show here that the enteric pathogen Salmonella Typhimurium can stimulate innate immune responses in cultured epithelial cells by mechanisms that do not involve receptors of the innate immune system. Instead, S. Typhimurium stimulates these responses by delivering through its type III secretion system the bacterial effector proteins SopE, SopE2, and SopB, which in a redundant fashion stimulate Rho-family GTPases leading to the activation of mitogen-activated protein (MAP) kinase and NF-kappaB signaling. These observations have implications for the understanding of the mechanisms by which Salmonella Typhimurium induces intestinal inflammation as well as other intestinal inflammatory pathologies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
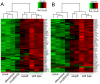
Figure 1. Salmonella Typhimurium type III secretion effector proteins induce innate immune responses in epithelial cells.
A Cluster analysis of the differentially expressed genes using the ratio of log2-transformed fold change in gene expression values (relative to uninfected cells) in cells infected with the indicated strains of S. Typhimurium. Genes and strains were clustered using unsupervised hierarchical clustering with complete linkage, and the data are visualized as row-normalized heat maps as indicated in the color scale. Each vertical row represents an independent experiment. Shown are genes that exhibited at least 4-fold change of expression (over uninfected controls) in all three experiments in cells infected with wild-type S. Typhimurium. The full data set are provided in Table S1. B Cluster analysis of differentially expressed genes which exhibited at least 2 fold change of expression (over uninfected controls) in all three experiments in cells infected with wild-type S. Typhimurium and that had been previously shown to be part of the pathogen response gene cluster . Genes and strains were clustered using unsupervised hierarchical clustering with complete linkage, and the data are visualized as row-normalized heat maps as indicated in the color scale.
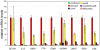
Figure 2. Quantitative RT-PCR analysis of the Salmonella Typhimurium-induced transcriptional response.
Cultured human epithelial cells were infected with different strains of S. Typhimurium (as indicated) and the levels of expression of selected genes were analyzed by qRT-PCR after reverse transcription of RNA samples extracted from cells infected with the indicated S. Typhimurium strains. The transcript levels were normalized to the levels of GAPDH in each sample, which were found to remain constant under any of the conditions used in the microarray experiments. Values are expressed as percentage of the fold change observed in cells infected with wild type (considered 100%) and represent the mean±s. e. m. of at least three independent experiments. Fold change for the different reporter genes after infection with wild type was as follows: SCCA1: 749; IL-8: 105; MIP2: 100; TTP: 52; EGR1: 89; IGFBP1: 11; LMO7: 54; GEM: 38; JUN: 42. The differences between the values of cells infected with wild type vs those infected with ΔinvA, ΔsopE/ΔSopE2/ΔsopB, or effectorless were statistically significant (_p_≥0.002, Student t test).
Figure 3. Stimulation of MAP kinases and NF-κB in epithelial cells infected with type III secretion mutants of Salmonella Typhimurium.
Cultured human epithelial cells were infected with different strains of S. Typhimurium and at different times after infection (as indicated), activation of the different kinases was evaluated by western immunoblot using antibodies directed to the phosphorylated (activated) form of the MAP kinases, or to the NF-κB inhibitor IκBα. Equal loading of the different samples was confirmed by re-probing the blots with antibodies directed to actin and/or to the different kinases.
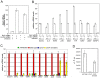
Figure 4. Salmonella Typhimurium stimulation of transcriptional responses in epithelial cells does not require the intracellular sensors.
A Depletion of Rip2 by RNAi. Cells transfected with siRNAs directed to Rip2 or with an irrelevant control siRNA were infected with wild-type S. Typhimurium and the levels of Rip2 in infected and uninfected cells were measured by qRT-PCR. The transcript levels were normalized to the levels of GAPDH. Values are expressed as percentage of the levels of Rip2 transcript in cells that had been treated with a control siRNA, which was considered 100%, and represent the mean±s. e. m. of at least three independent determinations. B Rip2 is not required for S. Typhimurium stimulation of gene expression in epithelial cells. Cells in which Rip2 had been depleted by RNAi were infected with the indicated strains of S. Typhimurium and the levels of expression of selected genes were analyzed by qRT-PCR after reverse transcription of RNA samples extracted from infected cells. The transcript levels were normalized to the levels of GAPDH. Values are expressed as percentage of the fold change observed in cells that had been treated with a control siRNA and infected with wild-type S. Typhimurium, which was considered 100%, and represent the mean±s. e. m. of at least three independent experiments. C S. Typhimurium internalized into epithelial cells through a heterologous internalization pathway does not stimulate the transcriptional responses induced by wild type. Cells were infected with a wild-type S. Typhimurium, an isogenic ΔinvA mutant, or the same mutant expressing Yersinia pseudotuberculosis invasin protein (p-invasin). The levels of expression of selected genes were analyzed by qRT-PCR after reverse transcription of RNA samples extracted from infected cells. The transcript levels were normalized to the levels of GAPDH. Values are expressed as percentage of the fold change observed in cells that had been infected with wild-type S. Typhimurium, which was considered 100%, and represent the mean±s. e. m. of at least three independent experiments. The differences between the values of cells infected with wild type vs those of uninfected cells or cells infected with the ΔinvA, or ΔinvA (p-invasin) strains were statistically significant (_p_≥0.001, student t test), except for the values corresponding to COX2 and IL-8 in cells infected with the ΔinvA (p-invasin) strain. D Levels of intracellular S. Typhimurium after internalization through the invasin-mediated pathway. Epithelial cells were infected with wild-type S. Typhimurium or the type III secretion-defective invA mutant expressing the Yersinia pseudotuberculosis invasin protein and the levels of intracellular bacteria were measured by the gentamicin protection assay as indicated in Materials and Methods. Results represent the colony forming units that resisted the gentamicin treatment due to their intracellular location and are the mean±standard deviation of three independent experiments.
Figure 5. Cdc42 is required for Salmonella Typhimurium stimulation of transcriptional responses in epithelial cells.
A Depletion of Cdc42 by RNAi. Cells transfected with an siRNA directed to Cdc42 or control cells transfected with an irrelevant construct, were infected with wild-type S. Typhimurium and the levels of Cdc42 in infected and uninfected cells were measured by qRT-PCR. The transcript levels were normalized to the levels of GAPDH. Values are expressed as percentage of the levels of Cdc42 transcript in cells that had been treated with a control siRNA and infected with wild-type S. Typhimurium, which was considered 100%, and represent the mean±s. e. m. of at least three independent determinations. B Cells in which Cdc42 had been depleted by RNAi were infected with the indicated strains of S. Typhimurium and the levels of expression of selected genes were analyzed by qRT-PCR after reverse transcription of RNA samples extracted from infected cells. The transcript levels were normalized to the levels of GAPDH. Values are expressed as percentage of the fold change observed in cells that had been treated with a control siRNA, which was considered 100%, and represent the mean±s. e. m. of at least three independent experiments. * : indicate that values are statistically significantly different (P<0.001, Student t test) from those of the control infected cells. C Henle-407 cells were co-transfected with an IL-8 transcription firefly-luciferase reporter plasmid along with a plasmid encoding renilla luciferase (to standardize transfection), and when indicated, with the indicated amount of a plasmid encoding SopE, a vector control, or a plasmid encoding a dominant negative mutant form of Cdc42 (Cdc42N17). The stimulation of IL-8 transcription in transfected cells was assayed by measuring the levels of firefly luciferase as indicated in Materials and Methods. Values represent fold induction in cells transfected with the SopE plasmid over the value of cells transfected with the plasmid vector alone and are the mean±standard deviation of three independent measurements. * : values statistically significant different from vector control (P<0.001, Student t test).
Figure 6. S. Typhimurium induces intestinal inflammation in Rip2-deficient mice.
A C57BL/6 or rip2−/− mice were treated with 20 mg of streptomycin, and 24 hs after antibiotic treatment, mice were either mock infected or infected orally with 108 of wild type S. typhimurium, or the isogenic Δ_invA_ (type III secretion defective) mutant. Forty-eight hours after infection, ceca were removed, fixed, embedded in paraffin, and tissue sections were stained with hematoxilin and eosin. Bar indicates 100 µm. Similar results were obtained in four independent animals for each group. B Mice were treated and infected with S. typhimurium as indicated above and bacterial loads in the ceca were determined 48 hs post-infection.
Similar articles
- Salmonella stimulates pro-inflammatory signalling through p21-activated kinases bypassing innate immune receptors.
Sun H, Kamanova J, Lara-Tejero M, Galán JE. Sun H, et al. Nat Microbiol. 2018 Oct;3(10):1122-1130. doi: 10.1038/s41564-018-0246-z. Epub 2018 Sep 17. Nat Microbiol. 2018. PMID: 30224799 Free PMC article. - Role of SPI-1 secreted effectors in acute bovine response to Salmonella enterica Serovar Typhimurium: a systems biology analysis approach.
Lawhon SD, Khare S, Rossetti CA, Everts RE, Galindo CL, Luciano SA, Figueiredo JF, Nunes JE, Gull T, Davidson GS, Drake KL, Garner HR, Lewin HA, Bäumler AJ, Adams LG. Lawhon SD, et al. PLoS One. 2011;6(11):e26869. doi: 10.1371/journal.pone.0026869. Epub 2011 Nov 11. PLoS One. 2011. PMID: 22096503 Free PMC article. - Flagellin acting via TLR5 is the major activator of key signaling pathways leading to NF-kappa B and proinflammatory gene program activation in intestinal epithelial cells.
Tallant T, Deb A, Kar N, Lupica J, de Veer MJ, DiDonato JA. Tallant T, et al. BMC Microbiol. 2004 Aug 23;4:33. doi: 10.1186/1471-2180-4-33. BMC Microbiol. 2004. PMID: 15324458 Free PMC article. - Characterization of effector proteins translocated via the SPI1 type III secretion system of Salmonella typhimurium.
Ehrbar K, Mirold S, Friebel A, Stender S, Hardt WD. Ehrbar K, et al. Int J Med Microbiol. 2002 Feb;291(6-7):479-85. doi: 10.1078/1438-4221-00156. Int J Med Microbiol. 2002. PMID: 11890547 Review. - Innate immune response to Salmonella typhimurium, a model enteric pathogen.
Broz P, Ohlson MB, Monack DM. Broz P, et al. Gut Microbes. 2012 Mar-Apr;3(2):62-70. doi: 10.4161/gmic.19141. Epub 2012 Mar 1. Gut Microbes. 2012. PMID: 22198618 Free PMC article. Review.
Cited by
- Consistent activation of the β-catenin pathway by Salmonella type-three secretion effector protein AvrA in chronically infected intestine.
Lu R, Liu X, Wu S, Xia Y, Zhang YG, Petrof EO, Claud EC, Sun J. Lu R, et al. Am J Physiol Gastrointest Liver Physiol. 2012 Nov 15;303(10):G1113-25. doi: 10.1152/ajpgi.00453.2011. Epub 2012 Sep 13. Am J Physiol Gastrointest Liver Physiol. 2012. PMID: 22982337 Free PMC article. - The Salmonella kinase SteC targets the MAP kinase MEK to regulate the host actin cytoskeleton.
Odendall C, Rolhion N, Förster A, Poh J, Lamont DJ, Liu M, Freemont PS, Catling AD, Holden DW. Odendall C, et al. Cell Host Microbe. 2012 Nov 15;12(5):657-68. doi: 10.1016/j.chom.2012.09.011. Cell Host Microbe. 2012. PMID: 23159055 Free PMC article. - The Major RNA-Binding Protein ProQ Impacts Virulence Gene Expression in Salmonella enterica Serovar Typhimurium.
Westermann AJ, Venturini E, Sellin ME, Förstner KU, Hardt WD, Vogel J. Westermann AJ, et al. mBio. 2019 Jan 2;10(1):e02504-18. doi: 10.1128/mBio.02504-18. mBio. 2019. PMID: 30602583 Free PMC article. - Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut.
Praveschotinunt P, Duraj-Thatte AM, Gelfat I, Bahl F, Chou DB, Joshi NS. Praveschotinunt P, et al. Nat Commun. 2019 Dec 6;10(1):5580. doi: 10.1038/s41467-019-13336-6. Nat Commun. 2019. PMID: 31811125 Free PMC article. - A Nonpyroptotic IFN-γ-Triggered Cell Death Mechanism in Nonphagocytic Cells Promotes Salmonella Clearance In Vivo.
Ingram JP, Tursi S, Zhang T, Guo W, Yin C, A Wynosky-Dolfi M, van der Heijden J, Cai KQ, Yamamoto M, Finlay BB, Brodsky IE, Grivennikov SI, Tükel Ç, Balachandran S. Ingram JP, et al. J Immunol. 2018 May 15;200(10):3626-3634. doi: 10.4049/jimmunol.1701386. Epub 2018 Apr 13. J Immunol. 2018. PMID: 29654208 Free PMC article.
References
- Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. - PubMed
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
- Medzhitov R. Toll-like receptors and innate immunity. Nature Rev Immunol 1: Nature Rev Immunol. 2001;1:135–145. - PubMed
- Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–382. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical