ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool - PubMed (original) (raw)
. 2010 Jul;38(Web Server issue):W462-8.
doi: 10.1093/nar/gkq319. Epub 2010 Apr 30.
Affiliations
- PMID: 20435679
- PMCID: PMC2896148
- DOI: 10.1093/nar/gkq319
ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool
Jeffry D Sander et al. Nucleic Acids Res. 2010 Jul.
Abstract
ZiFiT (Zinc Finger Targeter) is a simple and intuitive web-based tool that provides an interface to identify potential binding sites for engineered zinc finger proteins (ZFPs) in user-supplied DNA sequences. In this updated version, ZiFiT identifies potential sites for ZFPs made by both the modular assembly and OPEN engineering methods. In addition, ZiFiT now integrates additional tools and resources including scoring schemes for modular assembly, an interface with the Zinc Finger Database (ZiFDB) of engineered ZFPs, and direct querying of NCBI BLAST servers for identifying potential off-target sites within a host genome. Taken together, these features facilitate design of ZFPs using reagents made available to the academic research community by the Zinc Finger Consortium. ZiFiT is freely available on the web without registration at http://bindr.gdcb.iastate.edu/ZiFiT/.
Figures
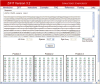
Figure 1.
ZiFiT Input Window. In this example the user has chosen to scan for ZFN sites using OPEN. The sequence has been entered in FASTA format, with exon sequences in uppercase and intron sequences in lowercase. Published sets of OPEN finger pools available through the Zinc Finger Consortium are checked by default.
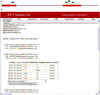
Figure 2.
ZiFiT Output Windows. (A) The Graphic Summary window (top) displays potential ZFN-binding sites (‘hits’) identified in the input sequence. Exons (denoted by uppercase characters in the input sequence) are displayed as thick red bars and introns as thin red lines. Hits are represented by short colored bars above the input sequence track; these serve as bookmarks that are linked to individual target sites in the Detailed Target List. ZFN hits are color-coded based on spacer size (5 bp = Blue; 6 bp = Green; 7 bp = Gold). (B) The Detailed Target List window (bottom) provides in-depth information about each hit. Hits are presented as double-stranded DNA sequences and labeled according to the FASTA description, spacer size, and index. Each hit can be expanded to reveal a reagent list for generating the corresponding ZFP. Hits can be sorted according to various criteria as detailed in the text.
Similar articles
- Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool.
Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Sander JD, et al. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W599-605. doi: 10.1093/nar/gkm349. Epub 2007 May 25. Nucleic Acids Res. 2007. PMID: 17526515 Free PMC article. - Zinc Finger Database (ZiFDB): a repository for information on C2H2 zinc fingers and engineered zinc-finger arrays.
Fu F, Sander JD, Maeder M, Thibodeau-Beganny S, Joung JK, Dobbs D, Miller L, Voytas DF. Fu F, et al. Nucleic Acids Res. 2009 Jan;37(Database issue):D279-83. doi: 10.1093/nar/gkn606. Epub 2008 Sep 23. Nucleic Acids Res. 2009. PMID: 18812396 Free PMC article. - Predicting success of oligomerized pool engineering (OPEN) for zinc finger target site sequences.
Sander JD, Reyon D, Maeder ML, Foley JE, Thibodeau-Beganny S, Li X, Regan MR, Dahlborg EJ, Goodwin MJ, Fu F, Voytas DF, Joung JK, Dobbs D. Sander JD, et al. BMC Bioinformatics. 2010 Nov 2;11:543. doi: 10.1186/1471-2105-11-543. BMC Bioinformatics. 2010. PMID: 21044337 Free PMC article. - Designer zinc finger proteins: tools for creating artificial DNA-binding functional proteins.
Dhanasekaran M, Negi S, Sugiura Y. Dhanasekaran M, et al. Acc Chem Res. 2006 Jan;39(1):45-52. doi: 10.1021/ar050158u. Acc Chem Res. 2006. PMID: 16411739 Review. - Zinc finger peptides for the regulation of gene expression.
Klug A. Klug A. J Mol Biol. 1999 Oct 22;293(2):215-8. doi: 10.1006/jmbi.1999.3007. J Mol Biol. 1999. PMID: 10529348 Review.
Cited by
- Efficient CRISPR/Cas9-mediated Targeted Mutagenesis in Populus in the First Generation.
Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K. Fan D, et al. Sci Rep. 2015 Jul 20;5:12217. doi: 10.1038/srep12217. Sci Rep. 2015. PMID: 26193631 Free PMC article. - EENdb: a database and knowledge base of ZFNs and TALENs for endonuclease engineering.
Xiao A, Wu Y, Yang Z, Hu Y, Wang W, Zhang Y, Kong L, Gao G, Zhu Z, Lin S, Zhang B. Xiao A, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D415-22. doi: 10.1093/nar/gks1144. Epub 2012 Nov 29. Nucleic Acids Res. 2013. PMID: 23203870 Free PMC article. - Heritable and precise zebrafish genome editing using a CRISPR-Cas system.
Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR. Hwang WY, et al. PLoS One. 2013 Jul 9;8(7):e68708. doi: 10.1371/journal.pone.0068708. Print 2013. PLoS One. 2013. PMID: 23874735 Free PMC article. - Comparison of CRISPR/Cas9 and TALENs on editing an integrated EGFP gene in the genome of HEK293FT cells.
He Z, Proudfoot C, Whitelaw CB, Lillico SG. He Z, et al. Springerplus. 2016 Jun 21;5(1):814. doi: 10.1186/s40064-016-2536-3. eCollection 2016. Springerplus. 2016. PMID: 27390654 Free PMC article. - Addiction associated N40D mu-opioid receptor variant modulates synaptic function in human neurons.
Halikere A, Popova D, Scarnati MS, Hamod A, Swerdel MR, Moore JC, Tischfield JA, Hart RP, Pang ZP. Halikere A, et al. Mol Psychiatry. 2020 Jul;25(7):1406-1419. doi: 10.1038/s41380-019-0507-0. Epub 2019 Sep 3. Mol Psychiatry. 2020. PMID: 31481756 Free PMC article.
References
- Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. - PubMed
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM069906/GM/NIGMS NIH HHS/United States
- R01 GM072621/GM/NIGMS NIH HHS/United States
- T32CA009216/CA/NCI NIH HHS/United States
- DP1 OD006862/OD/NIH HHS/United States
- R01 GM088040/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials