The roles of Drosophila cyclins A and B in mitotic control - PubMed (original) (raw)
Comparative Study
The roles of Drosophila cyclins A and B in mitotic control
C F Lehner et al. Cell. 1990.
Abstract
We have cloned, sequenced, and characterized the expression of a Drosophila cyclin B gene. The independent evolutionary conservation of A- and B-type cyclins implies that they have distinct roles. Indeed, in mutant embryos deficient in cyclin A, cells that accumulate only cyclin B do not enter mitosis. Thus, in vivo, cyclin B is not sufficient for mitosis. Furthermore, we find that the two cyclins are coexpressed in all proliferating cells throughout development. Though lacking a formal demonstration that cyclin B is essential as it is in other organisms, we propose that each of these proteins fulfills a distinct and essential role in the cell cycle.
Figures
Figure 1. Nucleotide Sequence and Predicted Amino Acid Sequence of a Drosophila Cyclin B cDNA
The nucleotide sequence of a Drosophila cyclin B cDNA (2691 bp) has a long open reading frame encoding a protein of 530 amino acids. The predicted amino acid sequence is shown above the nucleotide sequence. The putative initiating ATG is underlined along with flanking bases matching the consensus sequence for translational start sites in Drosophila (Cavener, 1987). The putative polyadenylation signal, followed by a poly(A) tail, is also underlined.
Figure 2. Amino Acid Sequence Comparison of Drosophila and Clam Cyclins
The amino acid sequences of the conserved cyclin domains of Drosophila cyclin B (D.B), clam cyclin B (C.B), Drosophila cyclin A (D.A), and clam cyclin A (C.A) are aligned. Gaps introduced for optimal alignment are indicated by dashes. Regions are boxed if the two A-type cyclins or the two B-type cyclins have similar amino acids (identical or conserved replacements using the following grouping: A,L,V,I,M; K,R; D,E; S,T; N,Q; Y,F). Positions similar in at least three sequences are marked with asterisks. Boldface indicates sequence identities in at least three of the four sequences. The clam sequences are according to Swenson et al. (1986) and Westendorf et al. (1989).
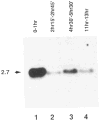
Figure 3. Cyclin B mRNA Levels during Embryonic Cell Cycle Progression
Total RNA from an equal number of early embryos during the syncytial cleavage divisions (lane 1), during interphase of Cycle 14 (lane 2), during cycle 15 and 16 (lane 3), and at a stage after cell divisions had become restricted mainly to the nervous system (lane 4) were probed on Northern blots with the cyclin B cDNA. The estimated size (kb) of the cyclin mRNA is indicated at left. Embryos were collected and aged at 25°C for the times indicated and staged under the microscope before isolation of RNA. Reprobing the filter with a probe specific for ribosomal protein rp49 (O’Connell and Rosbash, 1984) revealed roughly equal signal intensities in all four lanes (not shown).
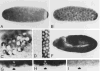
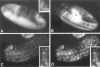
Figure 4. Localization of Cyclin B mRNA during Drosophila Embryogenesis
Cyclin B mRNA was localized by a color reaction after in situ hybridization with a digoxigeninylated cyclin B cDNA probe and incubation with alkaline phosphatase–conjugated anti-digoxigenin antibodies. Early embryos were staged after double labeling with the DNA stain Hoechst 33528. The DNA labeling is shown only in (C), which shows a combination of fluorescence and bright-field images. (A)Embryo before nuclear migration (cycle 3). Maximal signal intensities were found in a cap at the posterior end. In addition, signal intensities increased gradually toward the anterior end. (B)Embryo during nuclear migration (cycle 8). Cyclin B mRNA migrated in close association with the nuclei toward the periphery, and the signal at the polar cap was included in the polar buds. (C)Pole cells (cycle 14). After formation of the pole cells, cyclin B mRNA appeared to be sequestered into cytoplasmic granules clustered around the nuclei. (D)Somatic cells (cycle 14). No signal was detected in somatic cells if the color reaction was terminated early, as in (A)–(D). (E) Somatic cells (cycle 14). A specific signal in the cytoplasm of somatic cells was detected after prolonged color reaction as shown in (E)–(I). (F) Embryo during cycle 15. Signal was observed throughout the embryo except in the amnioserosa (labeled A). (G) Region from an embryo after cycle 16. No signal was detected in epidermal cells (between arrows) that had completed their developmental division program. Signal was present, however, in the nervous system (labeled N) where cell divisions continue. (H) Same region as in (G) but from an embryo homozygous for a null mutation in the cyclin A gene (neo114). As in wild-type embryos (G), cyclin B transcripts were no longer detected in epidermal cells during germband retraction. Mutant embryos were identified on the basis of their reduced nuclear densities as revealed by double labeling with the DNA stain Hoechst 33528 (not shown; see Lehner and O’Farrell, 1989). (I) Same as in (H) but hybridized with a cyclin A cDNA probe demonstrating the lack of zygotic cyclin A expression in homozygous neo114 embryos. In all panels except (C), anterior is left and ventral down. (C) shows the pole cells at the posterior end.
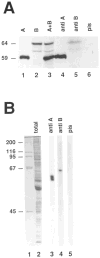
Figure 5. Characterization of Anti-Cyclin Antibodies
Immunoprecipitation experiments. Radiolabeled cyclin A (lane 1) and cyclin B (lane 2) were obtained after in vitro transcription–translation reactions. The two proteins were mixed (lane 3), and immunoprecipitations were done using affinity–purified rabbit antibodies against cyclin A (lane 4), a mouse antiserum against cyclin B (lane 5), or the corresponding mouse preimmune serum (lane 6). Proteins were resolved on a 7.5% SDS–polyacrylamide gel and visualized by autoradiography. Only the region containing cyclin A and cyclin B is shown. The estimated sizes (kd) of cyclin A and cyclin B are indicated at left. Immunoblotting experiments. Total protein extracts from 0–1 hr Drosophila embryos were resolved on a 10% SDS–polyacrylamide gel and stained with Coomassie blue (lane 2) or transferred to nitrocellulose (lanes 3–5) and probed with anti-cyclin A antibodies (lane 3), the mouse antiserum against cyclin B (lane 4), or the corresponding mouse preimmune serum (lane 5). Sizes (kd) of the marker proteins in lane 1 are indicated at left.
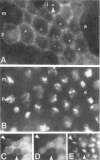
Figure 6. Intracellular Localization of Cyclin B during Interphase and Mitosis
Embryos were labeled with the antiserum against cyclin B(A) and with the DNA stain Hoechst 33258 (B). During interphase (i), the cytoplasm and intranuclear dots (arrowhead) were stained. In prophase (p), staining concentrated in the region of the condensing chromatin. Labeling became weak late in metaphase (m) and was undetectable in anaphase (a). In double-labeling experiments using rabbit anti-cyclin A(C), mouse anti-cyclin B antibodies (D), and Hoechst 33258 (E), cyclin A was undetectable in late metaphase cells (arrowheads) in which cyclin B was still detected. Primary antibodies were detected with rhodamine-conjugated (A, D) or fluorescein-conjugated (C) secondary antibodies.
Figure 7. Cyclin B Distribution during Embryogenesis
Embryos were stained with the antiserum against cyclin B followed by rhodamine-conjugated secondary antibodies (A, B, D). Double labeling with anti-cyclin A antibodies and fluorescein-conjugated secondary antibodies is shown in (C) and in the inset in (A). Embryos at the following stages are shown: (A) Onset of mitosis 14. Uniform cyclin B labeling is present in the cytoplasm. The same ratio between labeling intensity in somatic cells and pole cells (indicated by arrows) is observed with both anti-cyclin B antibodies and anti-cyclin A antibodies (inset). (B) Late in mitosis 14. Most cells have completed mitosis 14 and are not labeled with anti-cyclin B antibodies. Cells in the neurogenic region (N), in the amnioserosa (A), and in the posterior midgut (P) have not yet entered mitosis 14 and are intensely labeled. (C, D) After mitosis 16. Most cells have completed their developmental program of cell divisions and are no longer labeled. Cells in the central and peripheral nervous system that undergo additional mitotic divisions are labeled. Double labeling with anti-cyclin A(C) and anti-cyclin B antibodies (D) indicates that the two cyclins are coexpressed. The insets show a region of the embryo at a higher magnification. In all panels, anterior is left, ventral down.
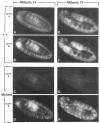
Figure 8. Cyclin Expression during Mitosis 15 and 16 in Wild-Type and Mutant Embryos
Wild-type embryos (A, B, E, F) and embryos homozygous for a null mutation in the cyclin A gene (C, D, G, H) during mitosis 15 (A–D) and mitosis 16 (E–H) are shown after double labeling with anti-cyclin A (A, C, E, G) and anti-cyclin B antibodies (B, D, F, H). For further explanations see text. In all panels anterior is left, ventral down.
Similar articles
- The A- and B-type cyclins of Drosophila are accumulated and destroyed in temporally distinct events that define separable phases of the G2-M transition.
Whitfield WG, Gonzalez C, Maldonado-Codina G, Glover DM. Whitfield WG, et al. EMBO J. 1990 Aug;9(8):2563-72. doi: 10.1002/j.1460-2075.1990.tb07437.x. EMBO J. 1990. PMID: 2142452 Free PMC article. - Expression and function of Drosophila cyclin A during embryonic cell cycle progression.
Lehner CF, O'Farrell PH. Lehner CF, et al. Cell. 1989 Mar 24;56(6):957-68. doi: 10.1016/0092-8674(89)90629-6. Cell. 1989. PMID: 2564316 Free PMC article. - Synergistic action of Drosophila cyclins A and B during the G2-M transition.
Knoblich JA, Lehner CF. Knoblich JA, et al. EMBO J. 1993 Jan;12(1):65-74. doi: 10.1002/j.1460-2075.1993.tb05632.x. EMBO J. 1993. PMID: 8428595 Free PMC article. - The A- and B-type cyclin associated cdc2 kinases in Xenopus turn on and off at different times in the cell cycle.
Minshull J, Golsteyn R, Hill CS, Hunt T. Minshull J, et al. EMBO J. 1990 Sep;9(9):2865-75. doi: 10.1002/j.1460-2075.1990.tb07476.x. EMBO J. 1990. PMID: 2143983 Free PMC article. - Pulling the string: cell cycle regulation during Drosophila development.
Lehner CF. Lehner CF. Semin Cell Biol. 1991 Aug;2(4):223-31. Semin Cell Biol. 1991. PMID: 1842341 Review.
Cited by
- Sgt1, a co-chaperone of Hsp90 stabilizes Polo and is required for centrosome organization.
Martins T, Maia AF, Steffensen S, Sunkel CE. Martins T, et al. EMBO J. 2009 Feb 4;28(3):234-47. doi: 10.1038/emboj.2008.283. Epub 2009 Jan 8. EMBO J. 2009. PMID: 19131964 Free PMC article. - Drosophila Cyclin B3 is required for female fertility and is dispensable for mitosis like Cyclin B.
Jacobs HW, Knoblich JA, Lehner CF. Jacobs HW, et al. Genes Dev. 1998 Dec 1;12(23):3741-51. doi: 10.1101/gad.12.23.3741. Genes Dev. 1998. PMID: 9851980 Free PMC article. - A role for cyclin D3 in the endomitotic cell cycle.
Zimmet JM, Ladd D, Jackson CW, Stenberg PE, Ravid K. Zimmet JM, et al. Mol Cell Biol. 1997 Dec;17(12):7248-59. doi: 10.1128/MCB.17.12.7248. Mol Cell Biol. 1997. PMID: 9372957 Free PMC article. - The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells.
Huang J, Raff JW. Huang J, et al. EMBO J. 1999 Apr 15;18(8):2184-95. doi: 10.1093/emboj/18.8.2184. EMBO J. 1999. PMID: 10205172 Free PMC article. - Regulation of Cyclin A protein in meiosis and early embryogenesis.
Vardy L, Pesin JA, Orr-Weaver TL. Vardy L, et al. Proc Natl Acad Sci U S A. 2009 Feb 10;106(6):1838-43. doi: 10.1073/pnas.0813237106. Epub 2009 Jan 30. Proc Natl Acad Sci U S A. 2009. PMID: 19181861 Free PMC article.
References
- Arion D, Meijer L, Brizuela L, Beach D. cdc2 is a component of the M phase–specific histone H1 kinase: evidence for identity with MPF. Cell. 1988;55:371–378. - PubMed
- Booher R, Alfa CE, Hyams JS, Beach D. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell. 1989;58:485–497. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous