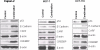Network modeling of MDM2 inhibitor-oxaliplatin combination reveals biological synergy in wt-p53 solid tumors - PubMed (original) (raw)
doi: 10.18632/oncotarget.269.
Sanjeev Banerjee, Shadan Ali, Zhiwei Wang, Bin Bao, Frances W J Beck, Main Maitah, Minsig Choi, Tony F Shields, Philip A Philip, Fazlul H Sarkar, Ramzi M Mohammad
Affiliations
- PMID: 21623005
- PMCID: PMC3248191
- DOI: 10.18632/oncotarget.269
Network modeling of MDM2 inhibitor-oxaliplatin combination reveals biological synergy in wt-p53 solid tumors
Asfar S Azmi et al. Oncotarget. 2011 May.
Abstract
Earlier we had shown that the MDM2 inhibitor (MI-219) belonging to the spiro-oxindole family can synergistically enhance the efficacy of platinum chemotherapeutics leading to 50% tumor free survival in a genetically complex pancreatic ductaladenocarcinoma (PDAC) xenograft model. In this report, we have taken a systems and network modeling approach in order to understand central mechanisms behind MI219-oxaliplatin synergy with validation in PDAC, colon and breast cancer cell lines. Microarray profiling of drug treatments (MI-219, oxaliplatin or their combination) in capan-2 cells reveal a similar unique set of gene alterations that is duplicated in other solid tumor cells. As single agent, MI-219 or oxaliplatin induced alterations in 48 and 761 genes respectively. The combination treatment resulted in 767 gene alterations with emergence of 286 synergy unique genes. Ingenuity network modeling of combination and synergy unique genes showed the crucial role of five key local networks CREB, CARF, EGR1, NF-kB and E-Cadherin. Compared to single agents the combination treatment super induced p53 and p21 confirming functional synergy. Further, the network signatures were validated at the protein level in all three cell lines. Individually silencing central nodes in these five hubsinterfered with MI-219-oxaliplatin activity confirming their critical role in aiding p53 mediated apoptotic response. We anticipate that our MI219-oxaliplatin network blueprints can be clinically translated in the rationale design and application of this unique therapeutic combination in a genetically pre-defined subset of patients.
Figures
Figure 1. MI-219 synergizes with Oxaliplatin leading to enhanced growth inhibition in wt-p53 cancer cells
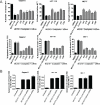
Figure 1 A) Three wt-p53 cancer cell lines Capan-2 representing PDAC; HCT116, colon cancer and MCF-7, a breast cancer cell line were seeded at a density of 3000 cells per well in 96 well plates. After 24 hrs the media was aspirated and cells were exposed to either MI-219 (0-30 μM) for Capan-2 or (0-15 μM for MCF-7 and HCT-116); oxaliplatin (0-30 μM) for Capan-2 or (0-15 μM for MCF-7 and HCT-116) or combination of MI-219 and Oxaliplatin (multiple concentrations in the ratio of 1:1 for isobologram analysis) for 32 hrs. At the end of the treatment period, 20 μL of MTT (Sigma St Louis USA) solution (5 mg/ml PBS) was added to each well and incubated at 37 °C for 2 hrs. After the incubation period was over, the media was aspirated and re-fed with 20 μL of DMSO per well following rapid mixing on a plate shaker. After 15 minutes of shaking the color developed was read at 595 nm using a ELISA Plate reader (TECAN Durham USA). Figure 1B) Isobologram analysis of MI-219-oxaliplatin combination treatment using CalcuSyn software a (CI<1 indicates synergy) and is observed in all cell lines.
Figure 2. MI-219-oxaliplatin induces synergistically enhanced apoptosis
[Upper Panel] Capan-2, MCF-7 and HCT-116 cells were grown in six well plates and treated with either MI-219 (15 μM) or oxaliplatin (15 μM) or the combination of MI-219 and Oxaliplatin (15 μM each) for 32 hrs. At the end of the treatment period, cells were trypsinized washed twice with PBS. The pelleted cells were tested for apoptosis using Annexin V FITC apoptosis assay according to the manufacturers guidelines (Biovision CA, USA Cat # K-101-100). [Lower Panel] Isobologram analysis of MI-219-oxaliplatin indicates synergy.
Figure 3. MI-219-Oxaliplatin suppresses colonogenic potential in different cancer cells
Cells were treated as described in Figure 1 and after the incubation was over, cells were plated in 100 mm petri plates in a density of 500-1000 cells depending on colonogenic potential. The plates were incubated in low CO2 days at 37°C in a 5% CO2/5% O2/90% N2 incubator. The colonies were stained with 2% crystal violet and counted. The surviving fraction was normalized to untreated control cells with respect to clonogenic efficiency.
Figure 4. MI-219-Oxaliplatin super induces p53 compared to single agent treatment
MCF-7 and Capan-2 cells grown in 100 mm petri plates were exposed to either (i) [Left to Right Lanes] vehicle control; (ii) MI-219 15 μM; (iii) Oxaliplatin 15 μM and (iv) combination of MI-219 and Oxaliplatin (15 μM each) for 32 hrs and protein was isolated as described in Methods section. Equal amounts of protein were analyzed for p53, MDM2 and p21 by Western Blotting. The membranes were re-probed with β-actin as loading control. Note in both cell lines the combination treatment lead to enhanced expression of p53, p21 and MDM2 in both cell lines.
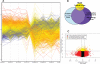
Figure 5. Microarray profiling of MI-219-Oxaliplatin treatment in Capan-2 PDAC cells
[A] Human Illumina HT-12 microarrays were performed on micro array grade RNA isolated from Capan-2 cells treated with either DMSO; MI-219 (15 μM); oxaliplatin (15 μM) or their combination for 16 and 32 hrs. Note: each treatment and time point has a unique set of gene expression profile (here color coded). All results represent at least triplicate samples except for oxaliplatin 32 hrs that was run in duplicate. [B] Biological Venn diagram showing each treatment induces unique set of gene changes (48 for MI-219 single agent treatment; 727 for oxaliplatin treatment; 767 for combination treatment and 286 genes as synergy unique genes. [C] Test for differential expression and variability indicates biomarker of response. Dataset of 24927 genes was analyzed in combination 32 hr vs. control 32 hrs. Differential gene expression: Combo vs Control (p = 0.01) Red = differentially expressed low variability Orange = differentially expressed high variability (potential biomarker of response). All values represent triplicate measurements with p value 0.01.
Figure 6. Combination synergy represents biologically meaningful process: Analysis of biologically activated functional pathways in combination treatment in (left panel) total combination genes vs
synergy unique genes (right panel). Note: enrichment of canonical functional pathways in combination datasets vs. restricted set of pathways in synergy unique datasets. Cutoff range was set at 1.7 in the ingenuity pathway analysis systems.
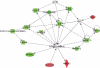
Figure 7. Network modeling and validation of MI-219 single agent treatment at different time points
[A] Ingenuity pathway analysis of primary events mediated by MI-219 single agent treatment at 16 hrs showing activation of NF-κB central hub. [B] Analysis of MI-219 single agent treatment mediated secondary events at 32 hrs showing de-regulated signaling and activation of secondary hub EGR1. Analysis presented in A and B are that of three biological replicates…
Figure 7. Network modeling and validation of MI-219 single agent treatment at different time points
[A] Ingenuity pathway analysis of primary events mediated by MI-219 single agent treatment at 16 hrs showing activation of NF-κB central hub. [B] Analysis of MI-219 single agent treatment mediated secondary events at 32 hrs showing de-regulated signaling and activation of secondary hub EGR1. Analysis presented in A and B are that of three biological replicates…
Figure 7C. Network modeling and validation of MI-219 single agent treatment at different time points
[C] Validation of primary and secondary gene changes at the functional level i.e. protein expression. Note activation of NF-κB p65 (p65) at 16 hrs and no significant changes in EGR1. However, at 32 hrs, p65 expression is lost and EGR1 hub is activated in all three cell lines. β-actin was used as loading control. Blots are representative of three independent experiments.
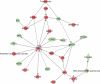
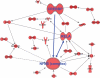
Figure 8. Ingenuity network analysis of total combination genes showing change in information flow in cells undergoing apoptosis: E-Cadherin represented by CDH1 anti-tumor module is activated in combination treatment
Red is indicative of genes going up and green depicts genes suppressed. All analysis presented are means of three biological replicates.
Figure 9. Global changes in gene homeostasis showing activation of NF-kB-CREB axis in synergy unique dataset
Triplicate datasets of the synergy unique genes were analyzed using ingenuity pathway analysis software and demonstrated activation of NF-kB module in tandem with CREB. Note all genes shown here are up-regulated.
Figure 10. Validation of network genes at the protein level: Capan-2, MCF-7 and HCT-116 cells were seeded in 100 mm petri dishes at the density of 1 million per plate in 10 ml media
Once the cells achieved 60-70% confluence, media was aspirated and replenished with fresh media containing either vehicle alone of MI-219+Oxaliplatin combination (15μM each). The cells were incubated for 32 hrs followed and at the end of the treatment period, cells were harvested, washed twice in PBS followed by extraction of protein for western blot analysis. Preparation of cellular lysates, protein concentration determination and SDS-PAGE analysis has been previously described (28). Equal amounts of protein were analyzed for p53, E-Cadherin, CARF and CREBBP by Western Blotting. The membranes were re-probed with β-actin as loading control. Note that the patterns of gene expression were similar in all three cell lines.
Figure 11. siRNA silencing of key hub genes abrogates MI-219-oxaliplatin efficacy
[A] Capan-2, MCF-7 and HCT-116 cells were grown in 96 well plates (for MTT Top Panel) and 6 well plates (for Annexin V FITC apoptosis Lower Panel). Semi confluent cells were treated with either DMSO; control siRNA or p53/CBP/EGR1/p16/p65 siRNAs for 5 hrs in serum free pencillin streptomycin free media for 5 hrs. After incubation, siRNA containing media was aspirated and cells were allowed to grow overnight in normal media. Following the overnight incubation, cells were exposed to MI-219-oxaliplatin combination for 32 hrs. Annexin V FITC apoptosis analysis was performed according to the manufacturer's guidelines (Biovision CA, USA Cat # K-101-100). [B] siRNA silencing of Cadherin under similar experimental conditions. All values [in A and B] represent Mean + S.D of triplicate experiment
Similar articles
- MDM2 inhibitor MI-319 in combination with cisplatin is an effective treatment for pancreatic cancer independent of p53 function.
Azmi AS, Aboukameel A, Banerjee S, Wang Z, Mohammad M, Wu J, Wang S, Yang D, Philip PA, Sarkar FH, Mohammad RM. Azmi AS, et al. Eur J Cancer. 2010 Apr;46(6):1122-31. doi: 10.1016/j.ejca.2010.01.015. Epub 2010 Feb 13. Eur J Cancer. 2010. PMID: 20156675 Free PMC article. - Reactivation of p53 by novel MDM2 inhibitors: implications for pancreatic cancer therapy.
Azmi AS, Philip PA, Aboukameel A, Wang Z, Banerjee S, Zafar SF, Goustin AS, Almhanna K, Yang D, Sarkar FH, Mohammad RM. Azmi AS, et al. Curr Cancer Drug Targets. 2010 May;10(3):319-31. doi: 10.2174/156800910791190229. Curr Cancer Drug Targets. 2010. PMID: 20370686 Free PMC article. - Reactivation of p53 by a specific MDM2 antagonist (MI-43) leads to p21-mediated cell cycle arrest and selective cell death in colon cancer.
Shangary S, Ding K, Qiu S, Nikolovska-Coleska Z, Bauer JA, Liu M, Wang G, Lu Y, McEachern D, Bernard D, Bradford CR, Carey TE, Wang S. Shangary S, et al. Mol Cancer Ther. 2008 Jun;7(6):1533-42. doi: 10.1158/1535-7163.MCT-08-0140. Mol Cancer Ther. 2008. PMID: 18566224 Free PMC article. - Spiro-oxindoles as a Promising Class of Small Molecule Inhibitors of p53-MDM2 Interaction Useful in Targeted Cancer Therapy.
Gupta AK, Bharadwaj M, Kumar A, Mehrotra R. Gupta AK, et al. Top Curr Chem (Cham). 2017 Feb;375(1):3. doi: 10.1007/s41061-016-0089-0. Epub 2016 Dec 9. Top Curr Chem (Cham). 2017. PMID: 27943171 Review. - Network perspectives on HDM2 inhibitor chemotherapy combinations.
Azmi AS, Beck FW, Sarkar FH, Mohammad RM. Azmi AS, et al. Curr Pharm Des. 2011;17(6):640-52. doi: 10.2174/138161211795222612. Curr Pharm Des. 2011. PMID: 21391913 Review.
Cited by
- Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascade inhibitors: how mutations can result in therapy resistance and how to overcome resistance.
McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Franklin RA, Montalto G, Cervello M, Libra M, Candido S, Malaponte G, Mazzarino MC, Fagone P, Nicoletti F, Bäsecke J, Mijatovic S, Maksimovic-Ivanic D, Milella M, Tafuri A, Chiarini F, Evangelisti C, Cocco L, Martelli AM. McCubrey JA, et al. Oncotarget. 2012 Oct;3(10):1068-111. doi: 10.18632/oncotarget.659. Oncotarget. 2012. PMID: 23085539 Free PMC article. Review. - Strategy to enhance the therapeutic effect of doxorubicin in human hepatocellular carcinoma by selenocystine, a synergistic agent that regulates the ROS-mediated signaling.
Fan C, Zheng W, Fu X, Li X, Wong YS, Chen T. Fan C, et al. Oncotarget. 2014 May 15;5(9):2853-63. doi: 10.18632/oncotarget.1854. Oncotarget. 2014. PMID: 24797310 Free PMC article. - Recent progress in genetics of aging, senescence and longevity: focusing on cancer-related genes.
Berman AE, Leontieva OV, Natarajan V, McCubrey JA, Demidenko ZN, Nikiforov MA. Berman AE, et al. Oncotarget. 2012 Dec;3(12):1522-32. doi: 10.18632/oncotarget.889. Oncotarget. 2012. PMID: 23455653 Free PMC article. Review. - Aberrant epigenetic grooming of miRNAs in pancreatic cancer: a systems biology perspective.
Azmi AS, Beck FW, Bao B, Mohammad RM, Sarkar FH. Azmi AS, et al. Epigenomics. 2011 Dec;3(6):747-59. doi: 10.2217/epi.11.97. Epigenomics. 2011. PMID: 22126293 Free PMC article. Review. - Inhibition of GSK-3β activity can result in drug and hormonal resistance and alter sensitivity to targeted therapy in MCF-7 breast cancer cells.
Sokolosky M, Chappell WH, Stadelman K, Abrams SL, Davis NM, Steelman LS, McCubrey JA. Sokolosky M, et al. Cell Cycle. 2014;13(5):820-33. doi: 10.4161/cc.27728. Epub 2014 Jan 9. Cell Cycle. 2014. PMID: 24407515 Free PMC article.
References
- Pujol A, Mosca R, Farres J, Aloy P. Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol Sci. 2010;31:115–123. - PubMed
- Klipp E, Wade RC, Kummer U. Biochemical network-based drug-target prediction. Curr Opin Biotechnol. 2010;21:511–516. - PubMed
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. - PubMed
- Stelzl U, Wanker EE. The value of high quality protein-protein interaction networks for systems biology. Curr Opin Chem Biol. 2006;10:551–558. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous