Schizosaccharomyces pombe minichromosome maintenance-binding protein (MCM-BP) antagonizes MCM helicase - PubMed (original) (raw)
Schizosaccharomyces pombe minichromosome maintenance-binding protein (MCM-BP) antagonizes MCM helicase
Lin Ding et al. J Biol Chem. 2011.
Abstract
The minichromosome maintenance (MCM) complex, a replicative helicase, is a heterohexamer essential for DNA duplication and genome stability. We identified Schizosaccharomyces pombe mcb1(+) (Mcm-binding protein 1), an apparent orthologue of the human MCM-binding protein that associates with a subset of MCM complex proteins. mcb1(+) is an essential gene. Deletion of mcb1(+) caused cell cycle arrest after several generations with a cdc phenotype and disrupted nuclear structure. Mcb1 is an abundant protein, constitutively present across the cell cycle. It is widely distributed in cytoplasm and nucleoplasm and bound to chromatin. Co-immunoprecipitation suggested that Mcb1 interacts robustly with Mcm3-7 but not Mcm2. Overproduction of Mcb1 disrupted the association of Mcm2 with other MCM proteins, resulting in inhibition of DNA replication, DNA damage, and activation of the checkpoint kinase Chk1. Thus, Mcb1 appears to antagonize the function of MCM helicase.
Figures
FIGURE 1.
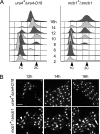
Spore germination of Δ_mcb1_ spores. A, spores prepared from wild-type ura4+/ura4D-18 (FY261x11) and heterozygous disruption mutant mcb1+/Δ_mcb1_::ura4+ (FY3747) were inoculated into medium lacking uracil at 32 °C. The populations were sampled every 2 h for 16 h and analyzed by flow cytometry to monitor S phase entry and DNA replication progression. B, photomicrographs of DAPI-stained spores after 12, 14, and 16 h at 32 °C. Arrowheads indicate cells with an abnormal nucleus. An arrow indicates cells with a normal nucleus. Scale bar, 10 μm.
FIGURE 2.
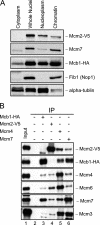
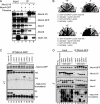
Mcb1 localization and its association to MCM. A, cellular fractions were prepared from mcb1HA mcm2V5 (FY4122) according to Fig. S6_A_. An equal volume (10 μl) of each fraction was separated by 8% or 15% SDS-PAGE and immunoblotted for Mcb1HA, Mcm2V5, Mcm7, α-tubulin (marker for cytosol and nucleoplasm), and Fib1 (Nop1; marker for chromatin). B, Mcb1 interacts with all other MCM proteins except Mcm2. Lysate was prepared from asynchronous mcb1HA mcm2V5 (FY4122) cells with B88 buffer. Twenty micrograms of soluble protein were loaded as input (lane 1). Identical amounts of lysate were precleared and immunoprecipitated (IP) with the antibodies shown. Ten microliters of immunoprecipitated sample (1:7, v/v) were used for each immunoblot. Samples were separated by 8% SDS-PAGE. Soluble lysate was immunoprecipitated with no antibody, anti-HA, anti-V5, anti-Mcm4, and anti-Mcm7 and immunoblotted for Mcm2V5, Mcb1HA, Mcm4, Mcm6, Mcm7, Mcm3, or Mcm5.
FIGURE 3.
Structure and function analysis of Mcb1. A, a schematic of Mcb1 truncations (1–10) made with a summary of their toxicity to wild-type cells, complementation of Δ_mcb1_, and interaction with Mcm4. B, wild-type cells (FY254) transformed with plasmids encoding Mcb1 truncations (1–10) were streaked on EMM-leucine plates with (+T; left) or without (−T; right) thiamine to repress or induce nmt1 promoter, respectively. aa, amino acids; ND, not determined.
FIGURE 4.
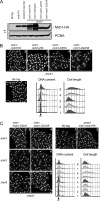
N-terminal deletion mutants (mcb1D2 and mcb1D22) are hypomorphic. A, wild-type (FY11), mcb1HA (FY4041), mcm2V5 mcb1HA (FY4122), Δ_mcb nmt1-mcb1HA_ (FY4596), Δ_mcb nmt1-mcb1gHA_ (FY5417), Δ_mcb nmt1-mcb1D2HA_ (FY5419), and Δ_mcb nmt1-mcb1D22HA_ (FY5421) cells were grown asynchronously in medium containing thiamine (+T). An equal number of cells were collected and alkaline lysed. An equal volume of total protein was loaded on an 8% SDS-polyacrylamide gel for separation and immunoblotted for Mcb1HA and proliferating cell nuclear antigen (PCNA) (a loading control). B, photomicrographs 1–5 of DAPI-stained asynchronous wild-type (FY11), Δ_mcb nmt1-mcb1HA_ (FY4596), Δ_mcb nmt1-mcb1gHA_ (FY5417), Δ_mcb nmt1-mcb1D2HA_ (FY5419) and Δ_mcb nmt1-mcb1D22HA_ (FY5421) cells. Scale bar, 10 μm. DNA content and cell length were monitored by flow cytometry. C, photomicrographs 1–8 of DAPI-stained wild-type (FY11); Δ_mcb nmt1-mcb1HA_ (FY4596); Δ_mcb nmt1-mcb1D2HA_ in a Δ_chk1_, Δ_cds1_, or Δ_rad3_ background (FY5499, -5501, or -5505); and Δ_mcb nmt1-mcb1D22HA_ in a Δ_chk1_, Δ_cds1_, or Δ_rad3_ background (FY5500, -5503, or -5506) cells. Scale bar, 10 μm. DNA content and cell length were monitored by flow cytometry.
FIGURE 5.
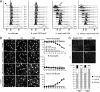
Overproduction of Mcb1 causes DNA damage. A, wild-type, negative control, and Mcb1 overproducing cells (a–d) at each time point were processed for flow cytometry analysis. Arrowheads indicate a starved 1C population. An arrow indicates a 2C population. Unfilled histograms outlined with dotted lines indicate FACS profiles of standard S. pombe cells processed at the same time. B, left, representative pictures of samples at each time point stained with DAPI. Arrowheads indicate short cells with abnormal nuclear morphology. Arrows indicate elongated cells with or without abnormal nuclear morphology. Scale bar, 10 μm. Right, quantification of cells with different nuclear morphology. Cells were counted for mononucleate, binucleate, and abnormal nucleus categories. C, Rad22-YFP foci formation in Mcb1-overproducing cells. rad22-YFP leu1-32::nmt1-mcb1HA-leu1+ cells (FY4591) were grown in −leucine with low thiamine to low OD, washed, and inoculated into −leucine with −thiamine (−T) or +thiamine (+T) medium. 14 and 16 h after thiamine removal, cells were harvested and washed twice in DAPI-containing medium. Left, representative photomicrographs of FY4591 at 14 h with or without thiamine. Scale bar, 10 μm. Cells were counted for Rad22-YFP foci (zero foci, one focus, and more than one focus). Right, quantification of cells counts averaged from two experiments.
FIGURE 6.
Chk1 is activated in Mcb1-overproducing cells. A, photomicrographs of DAPI-stained mcb1HA (FY4041), nmt1-mcb1HA (FY4594), Δ_cds1 nmt1-mcb1HA_ (FY4734), Δ_chk1 nmt1-mcb1HA_ (FY4736), Δ_cds1_ Δ_chk1 nmt1-mcb1HA_ (FY4739), and Δ_rad3 nmt1-mcb1HA_ (FY4740) cells 0 and 16 h after inoculation into −thiamine (−T) medium. Scale bar, 10 μm. B, chk1HA (FY4610) cells were transformed with plasmid expressing Mcb1V5 (pLD18) or empty vector (pSLF972). An equal number of cells were collected at the indicated time points after inoculation into −thiamine medium and alkaline lysed. An equal volume of protein was loaded on an SDS-polyacrylamide gel for separation and immunoblotted for Chk1HA, Mcb1V5, and α-tubulin (a loading control). Lane 1, total lysate of untreated chk1HA cells; lanes 2 and 3, total lysates of chk1HA cells treated with 0.1% methyl methanesulfonate (MMS) for 4 and 1 h (phosphorylated Chk1HA migrates slower), respectively; lanes 4-8, total lysates of pLD18-transformed chk1HA cells from different time points after thiamine removal; and lanes 9-13, total lysates of empty vector (pSLF972)-transformed chk1HA cells from different time points after thiamine removal.
FIGURE 7.
Overproduction of Mcb1 causes dissociation of Mcm2 from other MCM proteins. A, nmt1-mcb1HA mcm2V5 mcm4GFP (FY4961) overnight culture grown in low thiamine was inoculated into −thiamine (−T) and +thiamine (+T) media and grown at 32 °C for 14 h. Cells were harvested and lysed in B88 buffer. Soluble lysates were immunoprecipitated (IP) with anti-GFP (lanes 3 and 4) and anti-V5 (lanes 5 and 6). Immunoprecipitated samples were separated by SDS-PAGE gel and blotted for Mcm2V5, Mcm4GFP, Mcm7, Mcm6, and Mcb1HA. B, wild-type (FY254), nmt1-mcm4HA (FY1602), and nmt1-mcm2HA (FY861) cells were transformed with plasmids overexpressing Mcb1 (pLD18 or pLD10) and empty vector (pSLF972 or pSGP72). Transformants (top, a–d, and bottom, a–d) were restreaked on −thiamine and +thiamine plates and incubated at 32 °C. C, mcm2V5 mcm4GFP cells carrying full-length mcb1 (FY4961 and FY5407) and mcb1 deletion mutants (FY5408–5415) at the leu1-32 locus were grown in thiamine-containing medium, then harvested, and lysed in B88 buffer. Soluble lysates were immunoprecipitated with anti-GFP. Immunoprecipitated samples were separated by 12% SDS-PAGE. We used an 8% gel to separate bigger Mcb1 truncations from IgG (bottom panel). We blotted for Mcm4GFP and Mcb1HA. The data are summarized in Fig. 3_A. D_, mcm2V5 mcm4GFP cells carrying six mcb1 deletion mutants at the leu1-32 locus (nmt1-mcb1D2HA (FY5408), nmt1-mcb1D5HA (FY5411), nmt1-mcb1D6HA (FY5412), nmt1-mcb1D78HA (FY5413), nmt1-mcb1D9HA (FY5414), and nmt1-mcb1D22HA (FY5415)) were grown in −thiamine medium, then harvested, and lysed in B88 buffer. Soluble lysates were immunoprecipitated with anti-GFP. Fifteen micrograms of soluble proteins (lanes 1-8) and immunoprecipitated samples (lanes 9-14) were separated by SDS-PAGE and blotted for Mcm4GFP, Mcm2V5, Mcm6, Mcm7, and Mcb1HA.
FIGURE 8.
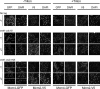
Overproduction of Mcb1 causes dissociation of chromatin-bound MCM proteins. nmt1-cdc18 mcm2V5 mcm4GFP (FY4958), nmt1-mcb1HA mcm2V5 mcm4GFP (FY4961), and wild-type (FY11) overnight cultures were grown in low thiamine, then inoculated into −thiamine (−T) or +thiamine (+T) medium, grown at 32 °C for 14 h, and harvested for an in situ chromatin binding assay. Mcb1HA and Mcm2V5 localization was detected in untreated cells and Triton-treated cells with specific antibodies. Scale bar, 10 μm.
Similar articles
- The fission yeast minichromosome maintenance (MCM)-binding protein (MCM-BP), Mcb1, regulates MCM function during prereplicative complex formation in DNA replication.
Santosa V, Martha S, Hirose N, Tanaka K. Santosa V, et al. J Biol Chem. 2013 Mar 8;288(10):6864-80. doi: 10.1074/jbc.M112.432393. Epub 2013 Jan 15. J Biol Chem. 2013. PMID: 23322785 Free PMC article. - Purification and functional inactivation of the fission yeast MCM(MCM-BP) complex.
Li JJ, Schnick J, Hayles J, MacNeill SA. Li JJ, et al. FEBS Lett. 2011 Dec 15;585(24):3850-5. doi: 10.1016/j.febslet.2011.10.033. Epub 2011 Oct 25. FEBS Lett. 2011. PMID: 22036784 - Identification and characterization of a novel component of the human minichromosome maintenance complex.
Sakwe AM, Nguyen T, Athanasopoulos V, Shire K, Frappier L. Sakwe AM, et al. Mol Cell Biol. 2007 Apr;27(8):3044-55. doi: 10.1128/MCB.02384-06. Epub 2007 Feb 12. Mol Cell Biol. 2007. PMID: 17296731 Free PMC article. - The MCM helicase: linking checkpoints to the replication fork.
Forsburg SL. Forsburg SL. Biochem Soc Trans. 2008 Feb;36(Pt 1):114-9. doi: 10.1042/BST0360114. Biochem Soc Trans. 2008. PMID: 18208397 Review. - Regulation of chromosome dynamics by Hsk1/Cdc7 kinase.
Matsumoto S, Masai H. Matsumoto S, et al. Biochem Soc Trans. 2013 Dec;41(6):1712-9. doi: 10.1042/BST20130217. Biochem Soc Trans. 2013. PMID: 24256280 Review.
Cited by
- The assembly of the MCM2-7 hetero-hexamer and its significance in DNA replication.
Hatoyama Y, Kanemaki MT. Hatoyama Y, et al. Biochem Soc Trans. 2023 Jun 28;51(3):1289-1295. doi: 10.1042/BST20221465. Biochem Soc Trans. 2023. PMID: 37145026 Free PMC article. Review. - MCMBP promotes the assembly of the MCM2-7 hetero-hexamer to ensure robust DNA replication in human cells.
Saito Y, Santosa V, Ishiguro KI, Kanemaki MT. Saito Y, et al. Elife. 2022 Apr 19;11:e77393. doi: 10.7554/eLife.77393. Elife. 2022. PMID: 35438632 Free PMC article. - Origin Firing Regulations to Control Genome Replication Timing.
Boos D, Ferreira P. Boos D, et al. Genes (Basel). 2019 Mar 6;10(3):199. doi: 10.3390/genes10030199. Genes (Basel). 2019. PMID: 30845782 Free PMC article. Review. - Genome-wide function of MCM-BP in Trypanosoma brucei DNA replication and transcription.
Kim HS. Kim HS. Nucleic Acids Res. 2019 Jan 25;47(2):634-647. doi: 10.1093/nar/gky1088. Nucleic Acids Res. 2019. PMID: 30407533 Free PMC article.
References
- Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., Oda M. (2010) Annu. Rev. Biochem. 79, 89–130 - PubMed
- Kelly T. J., Nurse P., Forsburg S. L. (1993) Cold Spring Harb. Symp. Quant. Biol. 58, 637–644 - PubMed
- Hartwell L. H., Weinert T. A. (1989) Science 246, 629–634 - PubMed
- Branzei D., Foiani M. (2008) Nat. Rev. Mol. Cell Biol. 9, 297–308 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous