Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes - PubMed (original) (raw)
Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes
Patrick T McGrath et al. Nature. 2011.
Abstract
Evolution can follow predictable genetic trajectories, indicating that discrete environmental shifts can select for reproducible genetic changes. Conspecific individuals are an important feature of an animal's environment, and a potential source of selective pressures. Here we show that adaptation of two Caenorhabditis species to growth at high density, a feature common to domestic environments, occurs by reproducible genetic changes to pheromone receptor genes. Chemical communication through pheromones that accumulate during high-density growth causes young nematode larvae to enter the long-lived but non-reproductive dauer stage. Two strains of Caenorhabditis elegans grown at high density have independently acquired multigenic resistance to pheromone-induced dauer formation. In each strain, resistance to the pheromone ascaroside C3 results from a deletion that disrupts the adjacent chemoreceptor genes serpentine receptor class g (srg)-36 and -37. Through misexpression experiments, we show that these genes encode redundant G-protein-coupled receptors for ascaroside C3. Multigenic resistance to dauer formation has also arisen in high-density cultures of a different nematode species, Caenorhabditis briggsae, resulting in part from deletion of an srg gene paralogous to srg-36 and srg-37. These results demonstrate rapid remodelling of the chemoreceptor repertoire as an adaptation to specific environments, and indicate that parallel changes to a common genetic substrate can affect life-history traits across species.
Figures
Figure 1
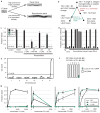
C. elegans cultivated in liquid are resistant to dauer pheromones. a, The developmental decision between reproductive growth and dauer larva formation is regulated by temperature, food, and population density. Population density is assessed by the release and sensation of ascarosides including C3, C5, C6, and C9. b, History of the C. elegans strains N2, LSJ1, LSJ2, and CC1 (see Methods). c, Dauer formation of N2, LSJ2, and CC1 in response to crude dauer pheromone or synthetic ascarosides. d, Dauer formation in response to synthetic C3 ascaroside. e, QTL mapping of C3 resistance. f, Schematic of near isogenic lines (NILs) with a small region from LSJ2 or CC1 introgressed into N2. g, Dauer formation in N2, LSJ2, and CX13249 strains. Error bars in all figures represent s.e.m.
Figure 2
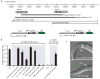
Resistance to C3 ascaroside is caused by deletion of two srg genes. a, Genomic region surrounding srg-36 and srg-37 on X, deletion breakpoints in LSJ2 and CC1 strains, fragments used for transgenic rescue, and design of bicistronic fusion genes. b, Transgenic rescue of dauer formation in response to C3 ascaroside. NIL strains used as recipients for rescue are shown in Figure 1f. ASI promoter was srg-47 (Figure S3), AFD promoter was gcy-8, ASE promoter was flp-6. c, Expression of GFP from bicistronic fusion genes for srg-36 and srg-37 in L1 larvae, showing predominant expression in ASI sensory neurons.
Figure 3
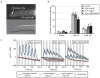
The srg genes encode ascaroside receptors. a, Localization of SRG-36∷GFP to ASI cilia (L4 animal). b, Ascaroside avoidance behaviours of animals with ectopic expression of srg-36, srg-37, or CBG24690 (shown in Figure 4) in the ASH nociceptive neurons. c, Ascaroside-induced Ca++ transients in ASH neurons that ectopically express C. elegans srg-36 or srg-37 or C. briggsae CBG24690 in ASH. Grey bars indicate the presence of C3 or C6 ascaroside, shading indicates s.e.m., n≥10 animals/condition. Ca++ was monitored using the genetically-encoded calcium sensor GCaMP3.0. Δ/F, percentage fluorescence change (baseline fluorescence = 100%).
Figure 4
Evolutionary conservation of srg function. a, Rescue of C. briggsae dauer formation in response to partially purified dauer pheromone by genomic fragments containing the CBG24690 gene. CX13431 is a near isogenic line containing the CBG24690 deletion from DR1690 introgressed into the AF16 background. b, Schematic of genes closely related to srg-36 and srg-37 from C. elegans, C. briggsae, and C. remanei (adapted from). c, CBG24690 genomic region from the AF16 C. briggsae reference strain, and location of a large deletion in the DR1690 C. briggsae strain that was cultivated for an extended period in liquid axenic media.
Similar articles
- Ascaroside expression in Caenorhabditis elegans is strongly dependent on diet and developmental stage.
Kaplan F, Srinivasan J, Mahanti P, Ajredini R, Durak O, Nimalendran R, Sternberg PW, Teal PE, Schroeder FC, Edison AS, Alborn HT. Kaplan F, et al. PLoS One. 2011 Mar 15;6(3):e17804. doi: 10.1371/journal.pone.0017804. PLoS One. 2011. PMID: 21423575 Free PMC article. - [Genetics and evolution of developmental plasticity in the nematode C. elegans: Environmental induction of the dauer stage].
Billard B, Gimond C, Braendle C. Billard B, et al. Biol Aujourdhui. 2020;214(1-2):45-53. doi: 10.1051/jbio/2020006. Epub 2020 Aug 10. Biol Aujourdhui. 2020. PMID: 32773029 Review. French. - A Forward Genetic Screen for Molecules Involved in Pheromone-Induced Dauer Formation in Caenorhabditis elegans.
Neal SJ, Park J, DiTirro D, Yoon J, Shibuya M, Choi W, Schroeder FC, Butcher RA, Kim K, Sengupta P. Neal SJ, et al. G3 (Bethesda). 2016 May 3;6(5):1475-87. doi: 10.1534/g3.115.026450. G3 (Bethesda). 2016. PMID: 26976437 Free PMC article. - Ascaroside activity in Caenorhabditis elegans is highly dependent on chemical structure.
Hollister KA, Conner ES, Zhang X, Spell M, Bernard GM, Patel P, de Carvalho AC, Butcher RA, Ragains JR. Hollister KA, et al. Bioorg Med Chem. 2013 Sep 15;21(18):5754-69. doi: 10.1016/j.bmc.2013.07.018. Epub 2013 Jul 18. Bioorg Med Chem. 2013. PMID: 23920482 Free PMC article. - Ascaroside Pheromones: Chemical Biology and Pleiotropic Neuronal Functions.
Park JY, Joo HJ, Park S, Paik YK. Park JY, et al. Int J Mol Sci. 2019 Aug 9;20(16):3898. doi: 10.3390/ijms20163898. Int J Mol Sci. 2019. PMID: 31405082 Free PMC article. Review.
Cited by
- Combinatorial chemistry in nematodes: modular assembly of primary metabolism-derived building blocks.
von Reuss SH, Schroeder FC. von Reuss SH, et al. Nat Prod Rep. 2015 Jul;32(7):994-1006. doi: 10.1039/c5np00042d. Nat Prod Rep. 2015. PMID: 26059053 Free PMC article. Review. - Interaction of structure-specific and promiscuous G-protein-coupled receptors mediates small-molecule signaling in Caenorhabditis elegans.
Park D, O'Doherty I, Somvanshi RK, Bethke A, Schroeder FC, Kumar U, Riddle DL. Park D, et al. Proc Natl Acad Sci U S A. 2012 Jun 19;109(25):9917-22. doi: 10.1073/pnas.1202216109. Epub 2012 Jun 4. Proc Natl Acad Sci U S A. 2012. PMID: 22665789 Free PMC article. - Selection and gene flow shape niche-associated variation in pheromone response.
Lee D, Zdraljevic S, Cook DE, Frézal L, Hsu JC, Sterken MG, Riksen JAG, Wang J, Kammenga JE, Braendle C, Félix MA, Schroeder FC, Andersen EC. Lee D, et al. Nat Ecol Evol. 2019 Oct;3(10):1455-1463. doi: 10.1038/s41559-019-0982-3. Epub 2019 Sep 23. Nat Ecol Evol. 2019. PMID: 31548647 Free PMC article. - A multi-parent recombinant inbred line population of C. elegans allows identification of novel QTLs for complex life history traits.
Snoek BL, Volkers RJM, Nijveen H, Petersen C, Dirksen P, Sterken MG, Nakad R, Riksen JAG, Rosenstiel P, Stastna JJ, Braeckman BP, Harvey SC, Schulenburg H, Kammenga JE. Snoek BL, et al. BMC Biol. 2019 Mar 12;17(1):24. doi: 10.1186/s12915-019-0642-8. BMC Biol. 2019. PMID: 30866929 Free PMC article. - Laboratory divergence of Methylobacterium extorquens AM1 through unintended domestication and past selection for antibiotic resistance.
Carroll SM, Xue KS, Marx CJ. Carroll SM, et al. BMC Microbiol. 2014 Jan 2;14:2. doi: 10.1186/1471-2180-14-2. BMC Microbiol. 2014. PMID: 24384040 Free PMC article.
References
- Protas ME, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–11. - PubMed
- Golden JW, Riddle DL. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol. 1984;102:368–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM07739/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- K99 GM092859/GM/NIGMS NIH HHS/United States
- R00 GM087533/GM/NIGMS NIH HHS/United States
- R00GM87533/GM/NIGMS NIH HHS/United States
- T32 GM007739/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous