An integrative approach to ortholog prediction for disease-focused and other functional studies - PubMed (original) (raw)
An integrative approach to ortholog prediction for disease-focused and other functional studies
Yanhui Hu et al. BMC Bioinformatics. 2011.
Abstract
Background: Mapping of orthologous genes among species serves an important role in functional genomics by allowing researchers to develop hypotheses about gene function in one species based on what is known about the functions of orthologs in other species. Several tools for predicting orthologous gene relationships are available. However, these tools can give different results and identification of predicted orthologs is not always straightforward.
Results: We report a simple but effective tool, the Drosophila RNAi Screening Center Integrative Ortholog Prediction Tool (DIOPT; http://www.flyrnai.org/diopt), for rapid identification of orthologs. DIOPT integrates existing approaches, facilitating rapid identification of orthologs among human, mouse, zebrafish, C. elegans, Drosophila, and S. cerevisiae. As compared to individual tools, DIOPT shows increased sensitivity with only a modest decrease in specificity. Moreover, the flexibility built into the DIOPT graphical user interface allows researchers with different goals to appropriately 'cast a wide net' or limit results to highest confidence predictions. DIOPT also displays protein and domain alignments, including percent amino acid identity, for predicted ortholog pairs. This helps users identify the most appropriate matches among multiple possible orthologs. To facilitate using model organisms for functional analysis of human disease-associated genes, we used DIOPT to predict high-confidence orthologs of disease genes in Online Mendelian Inheritance in Man (OMIM) and genes in genome-wide association study (GWAS) data sets. The results are accessible through the DIOPT diseases and traits query tool (DIOPT-DIST; http://www.flyrnai.org/diopt-dist).
Conclusions: DIOPT and DIOPT-DIST are useful resources for researchers working with model organisms, especially those who are interested in exploiting model organisms such as Drosophila to study the functions of human disease genes.
Figures
Figure 1
DIOPT facilitates identification of predicted orthologs based on various approaches, with flexible inputs and outputs. The DIOPT graphical user interface facilitates input of small or large gene or protein lists and is compatible with one or more of a variety of identifiers. Three filters are available: two of them exclude relationships that are predicted by < 2 or < 3 tools unless the only match score is equal to or lower than the threshold. The third one and most stringent filter limits outputs to the best-matching ortholog(s) per gene, as judged by the number of tools supporting the prediction. DIOPT provides a score based on the number of tools supporting the prediction and a weighted score based on the average of GO semantic similarity of orthologous pairs predicted by each tool using high quality GO functional annotation. In addition, DIOPT also provides the option to view the original score of each tool and a protein-protein alignment based on an updated proteome annotation (RefSeq release 44). Percent protein identity is calculated for both the overall alignment and domain regions.
Figure 2
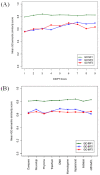
Assessing the correlation of functional relatedness and DIOPT score with gene ontology molecular function (GO MF) annotations. GO MF1, all GO annotations of molecular function. GO MF2, the subset of GO annotations of molecular function supported either by experimental data or author/curator statement. GO MF3, the subset of GO annotation of molecular function supported by experimental data. 2a. Correlation of functional relatedness and DIOPT score with GO MF annotations. 2b. Comparison of the functional relatedness of orthologous genes predicted by individual tools.
Figure 3
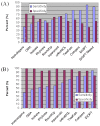
Comparing sensitivity and specificity among the nine individual ortholog prediction tools and the integrative tool DIOPT. Comparing sensitivity and specificity using a manually assembled reference set (3a), or using the KOG human-specific gene set (3b). Sensitivity is defined as the percent of manually assembled pairs that can be identified by each tool versus all the manually assembled orthologous pairs (same for Figures 3a and 3b). In 3a, specificity is defined as the percent of manually assembled pairs that can be identified by each tool versus all the orthologous pairs identified by each tool if queried with either the Drosophila or the human genes from the test set. In 3b, specificity is defined as the percent of putative human-specific genes that do not have fly ortholog versus all the human specific genes from KOG list.
Figure 4
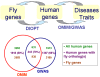
Identification of predicted disease gene orthologs in model organisms. 4a. Strategy for identifying disease genes in model organisms. 4b. Summary of disease genes from OMIM and GWAS and predicted orthologous relationships for Drosophila.
Figure 5
User Interface for the disease and trait query tool DIOPT-DIST. Users can query with a list of genes from model organisms, and a list of human orthologs along with their disease annotations from OMIM/GWAS will be retrieved. Alternatively, users can query with a disease term, disease category or OMIM IDs (OMIM IDs for disease phenotype and/or gene/locus), and a list of human genes as well as their corresponding orthologs in a model organism will be retrieved. The DIOPT score is displayed on the results page and is hyper-linked to detailed information including a protein alignment, the original tool scores, and the weighted DIOPT score.
Similar articles
- Prediction and enrichment analyses of the Homo sapiens-Drosophila melanogaster COPD-related orthologs: potential for modeling of human COPD genomic responses with the fruit fly.
Rouka E, Gourgoulianni N, Lüpold S, Hatzoglou C, Gourgoulianis KI, Zarogiannis SG. Rouka E, et al. Am J Physiol Regul Integr Comp Physiol. 2022 Jan 1;322(1):R77-R82. doi: 10.1152/ajpregu.00092.2021. Epub 2021 Dec 8. Am J Physiol Regul Integr Comp Physiol. 2022. PMID: 34877887 - Automatic clustering of orthologs and in-paralogs from pairwise species comparisons.
Remm M, Storm CE, Sonnhammer EL. Remm M, et al. J Mol Biol. 2001 Dec 14;314(5):1041-52. doi: 10.1006/jmbi.2000.5197. J Mol Biol. 2001. PMID: 11743721 - Paralog Explorer: A resource for mining information about paralogs in common research organisms.
Hu Y, Ewen-Campen B, Comjean A, Rodiger J, Mohr SE, Perrimon N. Hu Y, et al. Comput Struct Biotechnol J. 2022 Nov 24;20:6570-6577. doi: 10.1016/j.csbj.2022.11.041. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36467589 Free PMC article. - The Drosophila septate junctions beyond barrier function: Review of the literature, prediction of human orthologs of the SJ-related proteins and identification of protein domain families.
Rouka E, Gourgoulianni N, Lüpold S, Hatzoglou C, Gourgoulianis K, Blanckenhorn WU, Zarogiannis SG. Rouka E, et al. Acta Physiol (Oxf). 2021 Jan;231(1):e13527. doi: 10.1111/apha.13527. Epub 2020 Aug 5. Acta Physiol (Oxf). 2021. PMID: 32603029 Review. - OrthoDisease: tracking disease gene orthologs across 100 species.
Forslund K, Schreiber F, Thanintorn N, Sonnhammer EL. Forslund K, et al. Brief Bioinform. 2011 Sep;12(5):463-73. doi: 10.1093/bib/bbr024. Epub 2011 May 12. Brief Bioinform. 2011. PMID: 21565935 Review.
Cited by
- PomBase: a Global Core Biodata Resource-growth, collaboration, and sustainability.
Rutherford KM, Lera-Ramírez M, Wood V. Rutherford KM, et al. Genetics. 2024 May 7;227(1):iyae007. doi: 10.1093/genetics/iyae007. Genetics. 2024. PMID: 38376816 Free PMC article. Review. - Protein complex-based analysis framework for high-throughput data sets.
Vinayagam A, Hu Y, Kulkarni M, Roesel C, Sopko R, Mohr SE, Perrimon N. Vinayagam A, et al. Sci Signal. 2013 Feb 26;6(264):rs5. doi: 10.1126/scisignal.2003629. Sci Signal. 2013. PMID: 23443684 Free PMC article. - TDP-43 proteinopathy alters the ribosome association of multiple mRNAs including the glypican Dally-like protein (Dlp)/GPC6.
Lehmkuhl EM, Loganathan S, Alsop E, Blythe AD, Kovalik T, Mortimore NP, Barrameda D, Kueth C, Eck RJ, Siddegowda BB, Joardar A, Ball H, Macias ME, Bowser R, Van Keuren-Jensen K, Zarnescu DC. Lehmkuhl EM, et al. Acta Neuropathol Commun. 2021 Mar 24;9(1):52. doi: 10.1186/s40478-021-01148-z. Acta Neuropathol Commun. 2021. PMID: 33762006 Free PMC article. - Enhancer modeling uncovers transcriptional signatures of individual cardiac cell states in Drosophila.
Busser BW, Haimovich J, Huang D, Ovcharenko I, Michelson AM. Busser BW, et al. Nucleic Acids Res. 2015 Feb 18;43(3):1726-39. doi: 10.1093/nar/gkv011. Epub 2015 Jan 21. Nucleic Acids Res. 2015. PMID: 25609699 Free PMC article. - Higher resolution pooled genome-wide CRISPR knockout screening in Drosophila cells using Integration and Anti-CRISPR (IntAC).
Viswanatha R, Entwisle S, Hu C, Reap K, Butnaru M, Mohr SE, Perrimon N. Viswanatha R, et al. bioRxiv [Preprint]. 2024 Sep 25:2024.09.19.613976. doi: 10.1101/2024.09.19.613976. bioRxiv. 2024. PMID: 39345359 Free PMC article. Preprint.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous