Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion - PubMed (original) (raw)
Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion
Teresa L M Thurston et al. Nature. 2012.
Abstract
Autophagy defends the mammalian cytosol against bacterial infection. Efficient pathogen engulfment is mediated by cargo-selecting autophagy adaptors that rely on unidentified pattern-recognition or danger receptors to label invading pathogens as autophagy cargo, typically by polyubiquitin coating. Here we show in human cells that galectin 8 (also known as LGALS8), a cytosolic lectin, is a danger receptor that restricts Salmonella proliferation. Galectin 8 monitors endosomal and lysosomal integrity and detects bacterial invasion by binding host glycans exposed on damaged Salmonella-containing vacuoles. By recruiting NDP52 (also known as CALCOCO2), galectin 8 activates antibacterial autophagy. Galectin-8-dependent recruitment of NDP52 to Salmonella-containing vesicles is transient and followed by ubiquitin-dependent NDP52 recruitment. Because galectin 8 also detects sterile damage to endosomes or lysosomes, as well as invasion by Listeria or Shigella, we suggest that galectin 8 serves as a versatile receptor for vesicle-damaging pathogens. Our results illustrate how cells deploy the danger receptor galectin 8 to combat infection by monitoring endosomal and lysosomal integrity on the basis of the specific lack of complex carbohydrates in the cytosol.
Figures
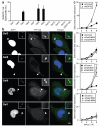
Figure 1. Galectin-8 responds to infection by S.Typhimurium and restricts bacterial proliferation
A-B) Analysis of HeLa cells stably expressing YFP fused to the indicated galectins and infected with S.Typhimurium for 1h. A) Percentage of bacteria coated by the indicated galectins. YFP-positive bacteria were counted by microscopy. Mean and s.d. of triplicate HeLa cultures, n>100 bacteria per coverslip. B) Confocal micrographs. Arrowheads, bacteria shown in insets. C) Kinetics of fold replication for S.Typhimurium in HeLa cells transfected with the indicated siRNAs. Bacteria were counted on the basis of their ability to form colonies on agar plates. Mean and s.d. of triplicate HeLa cultures and duplicate colony counts. siRNAs are further characterized in Supplementary Figure 2a-c. ** P<0.01, Student’s t-test. Scale bar 10 μm.
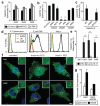
Figure 2. Galectin-8 binds NDP52
A) LUMIER binding assay: Normalized ratio between luciferase activity bound to beads and present in lysates. Lysates of 293ET cells expressing NDP52, p62, or Optineurin each fused to luciferase, and the indicated Flag-tagged galectins were incubated with anti-Flag beads. Flag-tagged proteins are further characterized in Supplementary Figure S4a. B) Lysates of 293ET cells, expressing Flag-tagged proteins as indicated, were immunoprecipitated with anti-Flag beads. Lysates and precipitates (IP) were blotted for the presence of Flag-tagged proteins and endogenous NDP52. C) Confocal images of HeLa cells infected with S.Typhimurium for 1h and stained with antisera against NDP52 and galectin-8. Arrowheads, bacteria shown in insets. D) Colocalization of NDP52 with galectin-positive bacteria in HeLa cells stably expressing YFP fused to the indicated galectins, infected with S.Typhimurium and stained with NDP52 antiserum 1h after infection. Mean and s.d. of duplicate HeLa cultures, n>100 bacteria per coverslip, representative of 2 independent experiments. Scale bar 10 μm.
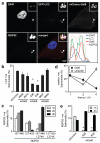
Figure 3. Galectin-8 is a danger receptor that senses cytosolic host glycans and recruits NDP52 to restrict Salmonella proliferation.
A) Percentage of S.Typhimurium coated by the indicated galectins. HeLa cells stably expressing YFP-tagged galectins were treated with the indicated siRNAs. YFP-positive bacteria were counted by microscopy at 1h post infection. siRNAs are further characterized in Supplementary Figure 2. B) Analysis of HeLa cells treated with the indicated siRNAs and stained with NDP52 antiserum. NDP52-positive bacteria were counted by microscopy 1h after infection with S.Typhimurium. siRNAs are further characterized in Supplementary Figure 2. C) Percentage of bacteria coated by the indicated galectin-8 alleles. HeLa cells stably expressing the indicated galectin-8 alleles fused to YFP were infected with S.Typhimurium. YFP-positive _S._Typhimurium were counted by microscopy at 75min p.i. D) Binding of galectin-8 to bacteria and HeLa cells. The indicated bacteria and HeLa cells were incubated with His-GST-ubiquitin, His-GST-galectin-8 or buffer as indicated, followed by murine anti-His antibody and PE-labelled anti-mouse serum. E) Percentage of S.Typhimurium coated by the indicated galectins. Wild type CHO cells and mutant Lec3.2.8.1 cells stably expressing YFP-tagged galectins were infected with _S._Typimurium. YFP-positive bacteria were counted by microscopy at 1h post infection. F) Confocal images of HeLa cells expressing the indicated YFP-tagged galectins. Cells were left untreated or were exposed to hypertonic conditions, with or without PEG as indicated, followed by hypotonic shock. G) Fold replication of S.Typhimurium in HeLa cells expressing the indicated galectin-3 variants and transfected with the indicated siRNAs. At 2 and 6h after infection, cells were lysed and bacteria counted on the basis of their ability to form colonies on agar plates. Galectin-3 proteins are further characterized in Supplementary Figure 4b. Mean and s.d. of duplicate coverslips (A-C,E) or triplicate HeLa cultures and duplicate colony counts (G). >100 bacteria counted per coverslip. Data are representative of at least two repeats. * P<0.05, Student’s t-test. Scale bar 10 μm
Figure 4. The anti-bacterial effect of galectin-8 is mediated by autophagy
A) Confocal micrograph of HeLa cells expressing GFP-LC3 and mCherry-galectin-8, stained for NDP52 1h after infection with S.Typhimurium. The lower right panel contains a fluorescence line scan along the yellow line in the merge inset. Arrowheads, bacteria shown in insets. B) Percentage of GFP-LC3 positive S.Typhimurium at 1h p.i. in HeLa cells treated with the indicated siRNAs. C) Percentage of bacteria positive for NDP52. HeLa cells expressing the indicated NDP52 variants fused to YFP were infected with _S._Typhimurium. D) Percentage of bacteria positive for the indicated markers. HeLa cells, either wild type or expressing YFP-galectin-8 as indicated, were infected with _S._Typhimurium. Ubiquitin was detected by antibody staining. E) HeLa cells treated with the indicated siRNAs were infected with _S._Typhimurium and stained for NDP52 at the indicated time points. Fluorescent bacteria were counted by microscopy at the indicated time points. Graphs, representing at least two independent repeats, show mean and s.d. of duplicate coverslips for which >200 bacteria were counted. * P<0.05, Student’s t-test, scale bar 10 μm.
Comment in
- Microbiology: A sweet way of sensing danger.
Huang J, Brumell JH. Huang J, et al. Nature. 2012 Feb 15;482(7385):316-7. doi: 10.1038/482316a. Nature. 2012. PMID: 22337047 No abstract available.
Similar articles
- Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy.
Li S, Wandel MP, Li F, Liu Z, He C, Wu J, Shi Y, Randow F. Li S, et al. Sci Signal. 2013 Feb 5;6(261):ra9. doi: 10.1126/scisignal.2003730. Sci Signal. 2013. PMID: 23386746 Free PMC article. - Microbiology: A sweet way of sensing danger.
Huang J, Brumell JH. Huang J, et al. Nature. 2012 Feb 15;482(7385):316-7. doi: 10.1038/482316a. Nature. 2012. PMID: 22337047 No abstract available. - Intracellular Salmonella induces aggrephagy of host endomembranes in persistent infections.
López-Montero N, Ramos-Marquès E, Risco C, García-Del Portillo F. López-Montero N, et al. Autophagy. 2016 Oct 2;12(10):1886-1901. doi: 10.1080/15548627.2016.1208888. Epub 2016 Aug 2. Autophagy. 2016. PMID: 27485662 Free PMC article. - Dual function of CALCOCO2/NDP52 during xenophagy.
Verlhac P, Viret C, Faure M. Verlhac P, et al. Autophagy. 2015;11(6):965-6. doi: 10.1080/15548627.2015.1046672. Autophagy. 2015. PMID: 25998689 Free PMC article. Review. - A Structural View of Xenophagy, a Battle between Host and Microbes.
Kwon DH, Song HK. Kwon DH, et al. Mol Cells. 2018 Jan 31;41(1):27-34. doi: 10.14348/molcells.2018.2274. Epub 2018 Jan 23. Mol Cells. 2018. PMID: 29370690 Free PMC article. Review.
Cited by
- "Repair Me if You Can": Membrane Damage, Response, and Control from the Viral Perspective.
Daussy CF, Wodrich H. Daussy CF, et al. Cells. 2020 Sep 7;9(9):2042. doi: 10.3390/cells9092042. Cells. 2020. PMID: 32906744 Free PMC article. Review. - Extracellular stimulation of VSIG4/complement receptor Ig suppresses intracellular bacterial infection by inducing autophagy.
Kim KH, Choi BK, Kim YH, Han C, Oh HS, Lee DG, Kwon BS. Kim KH, et al. Autophagy. 2016 Sep;12(9):1647-59. doi: 10.1080/15548627.2016.1196314. Epub 2016 Jul 20. Autophagy. 2016. PMID: 27440002 Free PMC article. - M. tuberculosis infection of human iPSC-derived macrophages reveals complex membrane dynamics during xenophagy evasion.
Bernard EM, Fearns A, Bussi C, Santucci P, Peddie CJ, Lai RJ, Collinson LM, Gutierrez MG. Bernard EM, et al. J Cell Sci. 2020 Nov 25;134(5):jcs252973. doi: 10.1242/jcs.252973. J Cell Sci. 2020. PMID: 32938685 Free PMC article. - Galectin-8N-Selective 4-Halophenylphthalazinone-Galactals Double π-Stack in a Unique Pocket.
van Klaveren S, Hassan M, Håkansson M, Johnsson RE, Larsson J, Jakopin Ž, Anderluh M, Leffler H, Tomašič T, Nilsson UJ. van Klaveren S, et al. ACS Med Chem Lett. 2024 Jul 22;15(8):1319-1324. doi: 10.1021/acsmedchemlett.4c00212. eCollection 2024 Aug 8. ACS Med Chem Lett. 2024. PMID: 39140038 Free PMC article. - Methicillin-resistant Staphylococcus aureus adaptation to human keratinocytes.
Soong G, Paulino F, Wachtel S, Parker D, Wickersham M, Zhang D, Brown A, Lauren C, Dowd M, West E, Horst B, Planet P, Prince A. Soong G, et al. mBio. 2015 Apr 21;6(2):e00289-15. doi: 10.1128/mBio.00289-15. mBio. 2015. PMID: 25900653 Free PMC article.
References
References for Methods
- Randow F, Sale JE. Retroviral transduction of DT40. Subcell Biochem. 2006;40:383–386. - PubMed
- Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. - PubMed
- Fitzgerald KA, et al. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. - PubMed
- Barrios-Rodiles M, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. - PubMed
References
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous