Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity - PubMed (original) (raw)
. 2012 Feb 1;482(7384):179-85.
doi: 10.1038/nature10809.
Eran Elinav, Chengcheng Jin, Liming Hao, Wajahat Z Mehal, Till Strowig, Christoph A Thaiss, Andrew L Kau, Stephanie C Eisenbarth, Michael J Jurczak, Joao-Paulo Camporez, Gerald I Shulman, Jeffrey I Gordon, Hal M Hoffman, Richard A Flavell
Affiliations
- PMID: 22297845
- PMCID: PMC3276682
- DOI: 10.1038/nature10809
Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity
Jorge Henao-Mejia et al. Nature. 2012.
Abstract
Non-alcoholic fatty liver disease (NAFLD) is the hepatic manifestation of metabolic syndrome and the leading cause of chronic liver disease in the Western world. Twenty per cent of NAFLD individuals develop chronic hepatic inflammation (non-alcoholic steatohepatitis, NASH) associated with cirrhosis, portal hypertension and hepatocellular carcinoma, yet the causes of progression from NAFLD to NASH remain obscure. Here, we show that the NLRP6 and NLRP3 inflammasomes and the effector protein IL-18 negatively regulate NAFLD/NASH progression, as well as multiple aspects of metabolic syndrome via modulation of the gut microbiota. Different mouse models reveal that inflammasome-deficiency-associated changes in the configuration of the gut microbiota are associated with exacerbated hepatic steatosis and inflammation through influx of TLR4 and TLR9 agonists into the portal circulation, leading to enhanced hepatic tumour-necrosis factor (TNF)-α expression that drives NASH progression. Furthermore, co-housing of inflammasome-deficient mice with wild-type mice results in exacerbation of hepatic steatosis and obesity. Thus, altered interactions between the gut microbiota and the host, produced by defective NLRP3 and NLRP6 inflammasome sensing, may govern the rate of progression of multiple metabolic syndrome-associated abnormalities, highlighting the central role of the microbiota in the pathogenesis of heretofore seemingly unrelated systemic auto-inflammatory and metabolic disorders.
Conflict of interest statement
The authors report no conflict of interest.
Figures
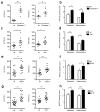
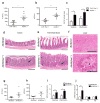
Figure 1. Increased severity of NASH in inflammasome-deficient mice
To induce NASH, mice were fed with MCDD for 24 d. Their serum ALT and AST activities measured and NAFLD histological activity scores were determined. (a–h) Comparison of ALT, AST, and NAFLD activity, plus histological scores for steatosis and inflammation between singly-housed wild-type (wt) mice and _caspase-1_−/− (a,b), _Asc_−/− (c,d), _Nlrp3_−/−(e,f), or _Il18_−/−(g,h). Data represent two independent experiments (n=7–19 mice/treatment group). Error bars represent the SEM of samples within a group. *p≤ 0.05, **p≤ 0.01, ***p≤ 0.001 (Student’s t test).
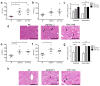
Figure 2. Increased severity of NASH in Asc and _Il18_-deficient mice is transmissible to co-housed wild-type animals
_Asc_−/− or _Il18_−/− mice and wt mice were co-housed for 4 weeks and then fed MCDD. (a–d) ALT (a), AST (b), NAFLD activity scores (c), and H&E-stained sections of livers (d) of singly-housed wt mice (wt), wt mice co-housed with _Asc_−/− (wt (_Asc_−/−)), and _Asc_−/− mice co-housed with wt (_Asc_−/−(wt)). (e–h) ALT (e), AST (f), NAFLD activity histological scores (g), and H&E-stained sections of livers (h) of wt, wt(_Il18_−/)−, and _Il18_−/−(wt). Data are representative of two independent experiments. Error bars represent SEM. Scale bars = 200 μm (d,h). *p≤ 0.05, **p≤ 0.01, ***p≤ 0.001.
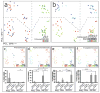
Figure 3. 16S rRNA sequencing demonstrates diet and co-housing associated changes in gut microbial ecology
(a) Principal Coordinates Analysis (PCoA) of Unweighted UniFrac distances of 16S rRNA sequences demonstrating clustering according to co-housing status on principal coordinate 1 (PC1). (b) PCoA of same plot as in (a) colored for experimental day. Mice were co-housed and fed a regular diet for the first 32 days of the experiment (two timepoints taken at day 20 and 32) before being switched to MCDD (sampled at days 39, 46 and 51 of the experiment). (c–f) PCoA and bargraphs of family level taxa Prevotellaceae, Porphyromonadaceae, Bacteroidaceae and Erysipelotrichaceae demonstrating diet-and microbiota-dependent differences in taxonomic representation. PCoA plots contain spheres represented a single fecal community colored according relative representation of the taxon (Blue represents relatively higher levels; red indicates lower levels). Bar graphs represent averaged taxonomic representation for singly or co-housed mouse while on either regular or MCD diet (n=8 for singly-housed wt, n=12 co-housed Asc_−/−(wt_) and wt(Asc_−/_−) animals;) * p < 0.05, ** p < 0.01, *** p < 0.001 by t-test after Bonferonni correction for multiple hypotheses. n.d. = not detected.
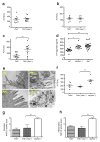
Figure 4. Increased severity of NASH in _Asc_-deficient and co-housed wild-type animals is mediated by TLR4, TLR9, and _TNF_-α
_Asc_−/− mice were co-housed with wt, Tnfα −/−, _Tlr4_−/−, _Tlr9_−/−, or _Tlr5_−/− mice for 4 weeks and then fed MCDD. (a–c) ALT levels of _Tlr4_−/−(_Asc_−/−) (a), _Tlr9_−/−(_Asc_−/−) (b), and _Tlr5_−/−(_Asc_−/−) (c) and their singly-housed counterparts. (d) TLR4 agonists in portal vein sera from MCDD-fed wt, wt(_Asc_−/−), and _Asc_−/− animals. (e) Transmission electron microscopy images of colon from wt and _Asc_−/−. (f–h) ALT (f), and NAFLD activity histological scores (g–h) of Tnfα −/−, wt(_Asc_−/−), and Tnfα −/− (_Asc_−/−). Data are representative of two independent experiments. Error bars represent SEM. *p≤ 0.05, **p≤ 0.01, ***p≤ 0.001.
Figure 5. Increased severity of NASH in _Asc_-deficient mice is transmissible to db/db by co-housing and is mediated by CCL5-induced intestinal inflammation
(a–c) ALT (a), AST (b), and NAFLD activity histological scores (c) of wt(_Asc_−/−) and _Ccl5_−/−(_Asc_−/−) mice. Data represents two independent experiments. (d–j) db/db mice were co-housed with wt or _Asc_−/− mice for 12 weeks. (d–f) Representative H&E-stained sections of colon (d), terminal ileum (e), and liver (f) from db/db(wt) and db/db(_Asc_−/−) mice fed a standard chow diet. Mucosal and crypt hyperplasia (arrow). Hepatocyte degeneration (arrowhead). Scale bars = 500 μm (d–e), Scale bars = 200 μm (f). (g–i) ALT (g), AST (h), and NAFLD activity scores (i) of db/db(wt), and db/db(Asc_−/−) mice. (j) Hepatic T_nfα, il6, and il1β mRNA levels. Error bars represent SEM. *p≤ 0.05, **p≤ 0.01, ***p≤ 0.001.
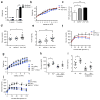
Figure 6. _Asc_-deficient mice develop increased obesity and loss of glycemic control on HFD
(a) Weight of db/db(wt) or (db/db(_Asc_−/−) at 3 weeks of age and at 12 weeks of co-housing. (b–f) _Asc_−/− and wt mice were co-housed for 4 weeks and then fed HFD. (b) Body weights. (c) NAFLD histological activity score. (d–e) Fasting plasma glucose and insulin after 11 weeks of HFD. (f) IPGTT after 12 weeks of HFD. (g–j) Mice were untreated, or treated orally with antibiotics, for 3 weeks prior to HFD feeding for 12 weeks. (g) Body weights. (h–i) Fasting plasma glucose and insulin levels after 8 weeks on a HFD. (j) IPGTT after 10 weeks of HFD. Error bars represent SEM. *p≤ 0.05, **p≤ 0.01, ***p≤ 0.001.
Comment in
- Gut microbiota influences liver disease.
Bird L. Bird L. Nat Rev Immunol. 2012 Feb 17;12(3):153. doi: 10.1038/nri3177. Nat Rev Immunol. 2012. PMID: 22343570 No abstract available. - Microbiota: Dysbiosis driven by inflammasome deficiency exacerbates hepatic steatosis and governs rate of NAFLD progression.
Wood NJ. Wood NJ. Nat Rev Gastroenterol Hepatol. 2012 Feb 21;9(3):123. doi: 10.1038/nrgastro.2012.21. Nat Rev Gastroenterol Hepatol. 2012. PMID: 22349167 No abstract available. - Is predisposition to NAFLD and obesity communicable?
Vijay-Kumar M, Gewirtz AT. Vijay-Kumar M, et al. Cell Metab. 2012 Apr 4;15(4):419-20. doi: 10.1016/j.cmet.2012.03.013. Cell Metab. 2012. PMID: 22482724 - Transmissible fatty liver disease.
Bromberg JS. Bromberg JS. Am J Transplant. 2012 May;12(5):1071. doi: 10.1111/j.1600-6143.2012.04095.x. Am J Transplant. 2012. PMID: 22537259 No abstract available. - Liver repercussions of defective gut surveillance.
Irvine KM, Schroder K, Powell EE. Irvine KM, et al. Hepatology. 2012 Sep;56(3):1174-7. doi: 10.1002/hep.25944. Hepatology. 2012. PMID: 22930647 No abstract available.
Similar articles
- Inhibition of NLRP3 inflammasome by thioredoxin-interacting protein in mouse Kupffer cells as a regulatory mechanism for non-alcoholic fatty liver disease development.
He K, Zhu X, Liu Y, Miao C, Wang T, Li P, Zhao L, Chen Y, Gong J, Cai C, Li J, Li S, Ruan XZ, Gong J. He K, et al. Oncotarget. 2017 Jun 6;8(23):37657-37672. doi: 10.18632/oncotarget.17489. Oncotarget. 2017. PMID: 28499273 Free PMC article. - Both bone marrow-derived and non-bone marrow-derived cells contribute to AIM2 and NLRP3 inflammasome activation in a MyD88-dependent manner in dietary steatohepatitis.
Csak T, Pillai A, Ganz M, Lippai D, Petrasek J, Park JK, Kodys K, Dolganiuc A, Kurt-Jones EA, Szabo G. Csak T, et al. Liver Int. 2014 Oct;34(9):1402-13. doi: 10.1111/liv.12537. Epub 2014 Apr 17. Liver Int. 2014. PMID: 24650018 Free PMC article. - Cardiolipin inhibitor ameliorates the non-alcoholic steatohepatitis through suppressing NLRP3 inflammasome activation.
Liu J, Wang T, He K, Xu M, Gong JP. Liu J, et al. Eur Rev Med Pharmacol Sci. 2019 Sep;23(18):8158-8167. doi: 10.26355/eurrev_201909_19036. Eur Rev Med Pharmacol Sci. 2019. PMID: 31599445 - Role of NLRP3 Inflammasome in the Progression of NAFLD to NASH.
Wan X, Xu C, Yu C, Li Y. Wan X, et al. Can J Gastroenterol Hepatol. 2016;2016:6489012. doi: 10.1155/2016/6489012. Epub 2016 May 4. Can J Gastroenterol Hepatol. 2016. PMID: 27446858 Free PMC article. Review. - NLRP3 inflammasome in hepatic diseases: A pharmacological target.
Ramos-Tovar E, Muriel P. Ramos-Tovar E, et al. Biochem Pharmacol. 2023 Nov;217:115861. doi: 10.1016/j.bcp.2023.115861. Epub 2023 Oct 18. Biochem Pharmacol. 2023. PMID: 37863329 Review.
Cited by
- NLRP7 and related inflammasome activating pattern recognition receptors and their function in host defense and disease.
Radian AD, de Almeida L, Dorfleutner A, Stehlik C. Radian AD, et al. Microbes Infect. 2013 Jul-Aug;15(8-9):630-9. doi: 10.1016/j.micinf.2013.04.001. Epub 2013 Apr 22. Microbes Infect. 2013. PMID: 23618810 Free PMC article. Review. - Short-Term Cohousing of Sick with Healthy or Treated Mice Alleviates the Inflammatory Response and Liver Damage.
Shabat Y, Lichtenstein Y, Ilan Y. Shabat Y, et al. Inflammation. 2021 Apr;44(2):518-525. doi: 10.1007/s10753-020-01348-0. Epub 2020 Sep 25. Inflammation. 2021. PMID: 32978699 - Discovering probiotic microorganisms: in vitro, in vivo, genetic and omics approaches.
Papadimitriou K, Zoumpopoulou G, Foligné B, Alexandraki V, Kazou M, Pot B, Tsakalidou E. Papadimitriou K, et al. Front Microbiol. 2015 Feb 17;6:58. doi: 10.3389/fmicb.2015.00058. eCollection 2015. Front Microbiol. 2015. PMID: 25741323 Free PMC article. Review. - Gut-central nervous system axis is a target for nutritional therapies.
Pimentel GD, Micheletti TO, Pace F, Rosa JC, Santos RV, Lira FS. Pimentel GD, et al. Nutr J. 2012 Apr 10;11:22. doi: 10.1186/1475-2891-11-22. Nutr J. 2012. PMID: 22490672 Free PMC article. Review.
References
- Sheth SG, Gordon FD, Chopra S. Nonalcoholic steatohepatitis. Ann Intern Med. 1997;126:137–145. - PubMed
- Ludwig J, Viggiano TR, McGill DB, Oh BJ. Nonalcoholic steatohepatitis: Mayo Clinic experiences with a hitherto unnamed disease. Mayo Clin Proc. 1980;55:434–438. - PubMed
- Marchesini G, et al. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology. 2003;37:917–923. - PubMed
- Caldwell SH, et al. Cryptogenic cirrhosis: clinical characterization and risk factors for underlying disease. Hepatology. 1999;29:664–669. - PubMed
- Shimada M, et al. Hepatocellular carcinoma in patients with non-alcoholic steatohepatitis. J Hepatol. 2002;37:154–160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R24 DK-085638/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U24 DK059635/DK/NIDDK NIH HHS/United States
- P30 DK-45735/DK/NIDDK NIH HHS/United States
- U24 DK-059635/DK/NIDDK NIH HHS/United States
- P30 DK045735-14/DK/NIDDK NIH HHS/United States
- T32 HL007974/HL/NHLBI NIH HHS/United States
- R01 DK-40936/DK/NIDDK NIH HHS/United States
- R01 DK076674/DK/NIDDK NIH HHS/United States
- R01 DK040936/DK/NIDDK NIH HHS/United States
- K08A1085038/PHS HHS/United States
- R01DK076674-01/DK/NIDDK NIH HHS/United States
- R24 DK085638/DK/NIDDK NIH HHS/United States
- P30 DK045735/DK/NIDDK NIH HHS/United States
- T32HL007974/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous