Altered transcriptome signature of phenotypically normal skin fibroblasts heterozygous for CDKN2A in familial melanoma: relevance to early intervention - PubMed (original) (raw)
Altered transcriptome signature of phenotypically normal skin fibroblasts heterozygous for CDKN2A in familial melanoma: relevance to early intervention
Meiyun Fan et al. Oncotarget. 2013 Jan.
Abstract
Familial melanoma (FM) is a dominantly heritable cancer that is associated with mutations in the tumor suppressor CDKN2A/p16. In FM, a single inherited "hit" occurs in every somatic cell, enabling interrogation of cultured normal skin fibroblasts (SFs) from FM gene carriers as surrogates for the cell of tumor origin, namely the melanocyte. We compared the gene expression profile of SFs from FM individuals with two distinct CDKN2A/p16 mutations (V126D-p16 and R87P-p16) with the gene expression profile of SFs from age-matched individuals without p16 mutations and with no family history of melanoma. We show an altered transcriptome signature in normal SFs bearing a single-hit inherited mutation in the CDKN2A/p16 gene, wherein some of these abnormal alterations recapitulate changes observed in the corresponding cancer. Significantly, the extent of the alterations is mutation-site specific with the R87P-p16 mutation being more disruptive than the V126D-p16 mutation. We also examined changes in gene expression after exposure to ultraviolet (UV) radiation to define potential early biomarkers triggered by sun exposure. UV treatment of SFs from FM families induces distinct alterations in genes related to cell cycle regulation and DNA damage responses that are also reported to be dysregulated in melanoma. Importantly, these changes were diametrically opposed to UV-induced changes in SF from normal controls. We posit that changes identified in the transcriptome of SF from FM mutation carriers represent early events critical for melanoma development. As such, they may serve as specific biomarkers of increased risk as well as molecular targets for personalized prevention strategies in high-risk populations.
Figures
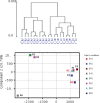
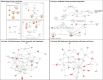
Figure 1. Hierarchical clustering and principal component analysis of gene expression data from SF cultures
Unsupervised hierarchical clustering (upper panel) and principal component analysis (lower panel) were performed on replicate cultures representing normal individuals with wild-type CDKN2A (N) or FM individuals with CDKN2A mutations (F) using normalized expression data of all probes.
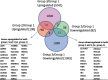
Figure 2. Differentially-expressed genes in familial SFs with distinct CDKN2A mutations
The inserted tables in the Venn Diagrams list the genes coordinately upregulated (left panel) or downregulated (right panel) in both Group 2 and Group 3 SFs in comparison to normal SFs (Group1).
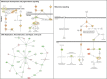
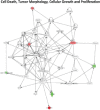
Figure 3. Signaling networks affected by the V126D-p16 mutation
Networks of differentially-expressed genes at baseline in familial SFs with the V126D-p16 mutation (Group 2) were generated using Ingenuity Pathway Analysis software. Genes upregulated or downregulated in Group 2 (vs. Group 1) were highlighted in red or green, respectively.
Figure 4. Signaling networks affected by the R87P-p16 mutation
Networks of differentially-expressed genes altered at baseline in familial SFs with a R87P-p16 mutation (Group 3) were generated using Ingenuity Pathway Analysis software. Genes upregulated or downregulated in Group 3 (vs. Group 1) were highlighted in red or green, respectively.
Figure 5. Induction of DNA damage by UV irradiation in SF cultures
SFs from a familial individual (F4) and a normal control individual (N1) were UV-irradiated for 20, 40 or 60 seconds, and then fixed in 3.7% formaldehyde at 2 hr after UV exposure. Double-stranded DNA breaks were visualized by γH2Ax immunofluorescence and nuclei were counterstained with DAPI.
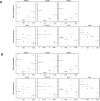
Figure 6. Hierarchical clustering and principal component analysis of gene expression data from SF cultures before and after UV irradiation
Unsupervised hierarchical clustering (upper panel) and principal component analysis (lower panel) were performed on gene expression data from un-irradiated and UV-irradiated SF cultures from the three cohorts using normalized expression data of all probes.
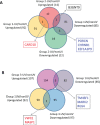
Figure 7. UV-affected genes in normal SFs and familial SFs with distinct CDKN2A mutations
Panel A. Venn diagrams of the UV-affected genes in normal SFs (Group 1) and familial SFs with a V126D-p16 mutation (Group 2). Four genes were coordinately regulated by UV irradiation (3 downregulated and 1 upregulated) in the two groups of SF cultures. Panel B. Venn diagrams of the UV-affected genes in normal SFs (Group 1) and familial SFs with a R87P-p16 mutation (Group 3). Five genes were oppositely regulated by UV irradiation in these two groups of SF cultures.
Figure 8. Signaling network of genes affected by UV in familial SFs with a V126D-p16 mutation
The gene network was generated using Ingenuity Pathway Analysis software. Genes upregulated or downregulated by UV in Group 2 were highlighted in red or green, respectively.
Figure 9. Signaling networks affected by UV in familial SFs with a R87P-p16 mutation
The gene networks were generated using Ingenuity Pathway Analysis software. Genes upregulated or downregulated by UV in Group 3 were highlighted in red or green, respectively.
Figure 10. Expression in normal skin and melanoma tissue of genes affected by UV in familial SFs with a R87P-p16 mutation
RNA was isolated from (Panel A) unirradiated and UV-irradiated SFs from Group 1 (normal) and Group 3 (a R87P-p16 mutation), or (Panel B) normal skin and melanoma tissue, and mRNA levels of the indicated genes were measured by qPCR, normalized to β-actin expression, and presented as means. The horizontal line shows the mean value for all members of each group of samples.
Similar articles
- Capturing the biological impact of CDKN2A and MC1R genes as an early predisposing event in melanoma and non melanoma skin cancer.
Puig-Butille JA, Escámez MJ, Garcia-Garcia F, Tell-Marti G, Fabra À, Martínez-Santamaría L, Badenas C, Aguilera P, Pevida M, Dopazo J, del Río M, Puig S. Puig-Butille JA, et al. Oncotarget. 2014 Mar 30;5(6):1439-51. doi: 10.18632/oncotarget.1444. Oncotarget. 2014. PMID: 24742402 Free PMC article. - A rare germline CDKN2A variant (47T>G; p16-L16R) predisposes carriers to pancreatic cancer by reducing cell cycle inhibition.
Horn IP, Marks DL, Koenig AN, Hogenson TL, Almada LL, Goldstein LE, Romecin Duran PA, Vera R, Vrabel AM, Cui G, Rabe KG, Bamlet WR, Mer G, Sicotte H, Zhang C, Li H, Petersen GM, Fernandez-Zapico ME. Horn IP, et al. J Biol Chem. 2021 Jan-Jun;296:100634. doi: 10.1016/j.jbc.2021.100634. Epub 2021 Apr 3. J Biol Chem. 2021. PMID: 33823155 Free PMC article. - Cloning and characterization of the CDKN2A and p19ARF genes from Monodelphis domestica.
Sherburn TE, Gale JM, Ley RD. Sherburn TE, et al. DNA Cell Biol. 1998 Nov;17(11):975-81. doi: 10.1089/dna.1998.17.975. DNA Cell Biol. 1998. PMID: 9839807 - Genetic Alterations in the INK4a/ARF Locus: Effects on Melanoma Development and Progression.
Ming Z, Lim SY, Rizos H. Ming Z, et al. Biomolecules. 2020 Oct 15;10(10):1447. doi: 10.3390/biom10101447. Biomolecules. 2020. PMID: 33076392 Free PMC article. Review. - Role of Heredity in Melanoma Susceptibility: A Primer for the Practicing Surgeon.
Abdo JF, Sharma A, Sharma R. Abdo JF, et al. Surg Clin North Am. 2020 Feb;100(1):13-28. doi: 10.1016/j.suc.2019.09.006. Epub 2019 Nov 1. Surg Clin North Am. 2020. PMID: 31753108 Review.
Cited by
- Capturing the biological impact of CDKN2A and MC1R genes as an early predisposing event in melanoma and non melanoma skin cancer.
Puig-Butille JA, Escámez MJ, Garcia-Garcia F, Tell-Marti G, Fabra À, Martínez-Santamaría L, Badenas C, Aguilera P, Pevida M, Dopazo J, del Río M, Puig S. Puig-Butille JA, et al. Oncotarget. 2014 Mar 30;5(6):1439-51. doi: 10.18632/oncotarget.1444. Oncotarget. 2014. PMID: 24742402 Free PMC article. - The pseudogenes of eukaryotic translation elongation factors (EEFs): Role in cancer and other human diseases.
Cristiano L. Cristiano L. Genes Dis. 2021 Apr 16;9(4):941-958. doi: 10.1016/j.gendis.2021.03.009. eCollection 2022 Jul. Genes Dis. 2021. PMID: 35685457 Free PMC article. Review. - ETHE1 overexpression promotes SIRT1 and PGC1α mediated aerobic glycolysis, oxidative phosphorylation, mitochondrial biogenesis and colorectal cancer.
Witherspoon M, Sandu D, Lu C, Wang K, Edwards R, Yeung A, Gelincik O, Manfredi G, Gross S, Kopelovich L, Lipkin S. Witherspoon M, et al. Oncotarget. 2019 Jun 18;10(40):4004-4017. doi: 10.18632/oncotarget.26958. eCollection 2019 Jun 18. Oncotarget. 2019. PMID: 31258845 Free PMC article. - Detection of Exosomal miRNAs in the Plasma of Melanoma Patients.
Pfeffer SR, Grossmann KF, Cassidy PB, Yang CH, Fan M, Kopelovich L, Leachman SA, Pfeffer LM. Pfeffer SR, et al. J Clin Med. 2015 Dec 17;4(12):2012-27. doi: 10.3390/jcm4121957. J Clin Med. 2015. PMID: 26694476 Free PMC article. - Tumor-suppressor Genes, Cell Cycle Regulatory Checkpoints, and the Skin.
Abreu Velez AM, Howard MS. Abreu Velez AM, et al. N Am J Med Sci. 2015 May;7(5):176-88. doi: 10.4103/1947-2714.157476. N Am J Med Sci. 2015. PMID: 26110128 Free PMC article. Review.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics. CA: a cancer journal for clinicians. 2012;62(1):10–29. - PubMed
- Narayanan DL, Saladi RN, Fox JL. Ultraviolet radiation and skin cancer. International journal of dermatology. 2010;49(9):978–986. - PubMed
- Kefford RF, Newton Bishop JA, Bergman W, Tucker MA. Counseling and DNA testing for individuals perceived to be genetically predisposed to melanoma: A consensus statement of the Melanoma Genetics Consortium. J Clin Oncol. 1999;17(10):3245–3251. - PubMed
- Piepkorn M. Melanoma genetics: an update with focus on the CDKN2A(p16)/ARF tumor suppressors. J Am Acad Dermatol. 2000;42(5 Pt 1):705–722. quiz 723-706. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous