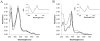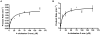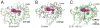A highly conserved mycobacterial cholesterol catabolic pathway - PubMed (original) (raw)
A highly conserved mycobacterial cholesterol catabolic pathway
Esther García-Fernández et al. Environ Microbiol. 2013 Aug.
Abstract
Degradation of the cholesterol side-chain in Mycobacterium tuberculosis is initiated by two cytochromes P450, CYP125A1 and CYP142A1, that sequentially oxidize C26 to the alcohol, aldehyde and acid metabolites. Here we report characterization of the homologous enzymes CYP125A3 and CYP142A2 from Mycobacterium smegmatis mc(2) 155. Heterologously expressed, purified CYP125A3 and CYP142A2 bound cholesterol, 4-cholesten-3-one, and antifungal azole drugs. CYP125A3 or CYP142A2 reconstituted with spinach ferredoxin and ferredoxin reductase efficiently hydroxylated 4-cholesten-3-one to the C-26 alcohol and subsequently to the acid. The X-ray structures of both substrate-free CYP125A3 and CYP142A2 and of cholest-4-en-3-one-bound CYP142A2 reveal significant differences in the substrate binding sites compared with the homologous M. tuberculosis proteins. Deletion only of cyp125A3 causes a reduction of both the alcohol and acid metabolites and a strong induction of cyp142 at the mRNA and protein levels, indicating that CYP142A2 serves as a functionally redundant back up enzyme for CYP125A3. In contrast to M. tuberculosis, the M. smegmatis Δcyp125Δcyp142 double mutant retains its ability to grow on cholesterol albeit with a diminished capacity, indicating an additional level of redundancy within its genome.
© 2013 John Wiley & Sons Ltd and Society for Applied Microbiology.
Figures
Fig. 1
Absolute Soret region absorption spectra of CYP125 (A) and CYP142 (B) in their resting (solid line), cholesterol-bound (dotted line) and econazole-bound (dashed line) forms. The protein concentration was always 3 μM. Cholesterol (in 10% of MβCD) and econazole (in methanol) were added at 50 μM. The insets show difference spectra generated by subtracting the spectra for the dithionite-treated enzymes from the ferrous-carbon monoxide complexes.
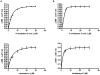
Fig. 2
Binding of cholest-4-en-3-one and cholesterol to CYP125A3 (A) and CYP142A2 (B). Ligand binding was monitored by the concentration dependent difference spectra observed in the Soret region for the substrates cholest-4-en-3-one (top) and cholesterol (bottom).
Fig. 3
CYP125A3 (A) and CYP142A2 (B) oxidation of cholest-4-en-3-one. Fitting results to the Michaelis-Menten kinetic equation (see Experimental Procedures) for substrate oxidation assays. Reactions were run for 5 to 20 min, the products were separated by HPLC, and the rate of 26-hydroxycholest-4-en-3-one formation was determined. Minor amounts of the aldehyde and acid products were observed in some reactions, but were not included in the analysis.
Fig. 4
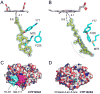
Overall structures of CYP125 and CYP142. (A) Cα traces for substrate-free CYP125A3 of M. smegmatis (green) and CYP125A1 of M. tuberculosis (magenta) are shown overlapped. The D108-K214 salt-bridge, missing from M. smegmatis, is shown in magenta sticks. Cholest-4-en-3-one (cyan sticks) is shown in the orientation observed in CYP125A1 of M. tuberculosis (PDB ID 2X5W). (B) Cα traces for substrate-bound CYP142A2 of M. smegmatis (green) and CYP142A1 of M. tuberculosis (magenta) are shown overlapped. The F-G-loop is in a more open conformation in the M. smegmatis structure. (C) Cα traces for substrate-bound CYP142A2 of M. smegmatis (green) and CYP125A1 of M. tuberculosis (grey) are shown overlapped. Highlighted in magenta and cyan are two amino acid inserts in CYP125 sequence that are missing from CYP142. Overlapped substrate molecules demonstrate the tilted position of cholest-4-en-3-one in CYP142A2 (green) compared to that of CYP125A1 (grey).
Fig. 5
Cholest-4-en-3-one binding. (A, B) Cholest-4-en-3-one in the CYP142A2 active site. The cholest-4-en-3-one molecule is shown in yellow sticks, with non-invariant residues clustered over the flat surface of the steroid ring system shown in cyan. Views in A and B differ by 90° rotation about the vertical axis. A fragment of the 2Fo-Fc electron density map (blue mesh) is contoured at 1.5σ. The distances between the heme iron and the carbon atoms of the branched methyl groups in cholest-4-en-3-one are in Angstroms. The smooth curvature of the ring system helps accommodate the bulkier side-chains in CYP124A2. (C, D) Surface representation of CYP125A1 (C) and CYP142A2 (D). Protein surfaces are colored by elements with carbon grey, oxygen red, nitrogen blue and sulfur yellow. (C) Amino acid inserts in the CYP125 sequences missing from CYP142 are mapped by magenta (102–111) and cyan (57–67), as indicated in the alignments in Fig. 6. (D) Cholest-4-en-3-one represented by VDW spheres exposes the carbonyl group (red) to the surface of CYP142A2. The exposed carbonyl group is surrounded largely by a hydrophobic area.
Fig. 6
Sequence alignments and analysis. The amino acid sequences for M. smegmatis (Ms) and M. tuberculosis (Mt) CYP125 and CYP142 are aligned using CLUSTALW (Thompson et al., 1994). UniProt database (
) accession numbers are provided for each protein sequence. Secondary structure elements are assigned based on 2YOO for CYP142A2 (top) and 2X5W for CYP125A1 (bottom). Residues interacting with cholest-4-en-3one within 5 A are highlighted in green. Cross-species differences are highlighted in yellow. Residue numbering on top is according to CYP142A2; residue numbering on bottom is according to CYP125A1. Amino acid inserts in the CYP125 sequences highlighted in cyan and pink are mapped to the CYP125A1 protein surface in Fig. 5 using the same colors.
Fig. 7
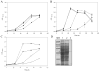
Growth analysis of mycobacterial mutant strains. Growth curves of WT (squares), _ΔCyp12_5 (circles), ΔCyp142 (triangles) and ΔCyp125ΔCyp142 (diamonds) on 1.8 mM of cholesterol (A) and 1.8 mM of cholest-4-en-3-one (B) as the sole carbon and energy source. Growth was monitored by measuring the absorbance at 600 nm. Data represent averages of duplicates and error bars indicate ±1 standard deviation.(C) Growth curves of ΔCyp125ΔCyp142 complemented with empty vector pMV261 (empty symbols) and pMVCyp125 (filled symbols) on cholesterol (squares) and cholest-4-en-3-one (circles). (D) Analysis of the production of CYP125A3 in the double mutant ΔCyp125ΔCyp142 complemented with plasmid pMVCyp125 by SDS-PAGE. MW, Broad range molecular markers (Bio-Rad); lane 1, protein extract of strain ΔCyp125ΔCyp142 carrying pMV261; lane 2, protein extract of strain ΔCyp125ΔCyp142 carrying pMVCyp125. As a control, purified CYP125A3 is loaded in lane 4. The narrow indicates the position of the CYP125A3 in the 10% of polyacrylamide gel.
Fig. 8
Analysis of the compounds present in supernatants of different M. smegmatis strains. The panels show LC-MS profiles of the 26-hydroxycholest-4-en-3-one (solid line) and cholest-4-en-3-one-26-oic acid (dashed line) versus time in wild-type and mutant strains grown on 1.8 mM cholest-4-en-3-one. Data represent compound peak area/internal standard peak area.
Fig. 9
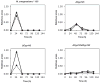
Analysis of endogenous expression of the CYP125A3 and CYP142A2 enzymes. (A) SDS/PAGE (12.5% gel) of protein extracts (40 μg) from mycobacterial mutants and M. smegmatis cytochromes (20 ng) The positions of wild-type and different mutant strains are indicated. MW, prestained protein molecular mass marker. (B) Western-blot analysis of endogenous production of CYP125 and CYP142 in the different mutant strains. (C) Differential expression of the cyp142 (black bars) and MSMEG_4829 (grey bars) genes in the wild-type, Δ_Cyp125_ and Δ_Cyp125_Δ_Cyp142_ strains cultured in cholesterol or glycerol. Transcription levels were measured using RTq-PCR as described in Experimental procedures. The values indicate the ratios of mRNA levels observed for strains growing on cholesterol relative to glycerol. Data represent averages of triplicates and error bars indicate ±1 standard deviation.
Similar articles
- Cytochrome P450 125A4, the Third Cholesterol C-26 Hydroxylase from Mycobacterium smegmatis.
Frank DJ, Waddling CA, La M, Ortiz de Montellano PR. Frank DJ, et al. Biochemistry. 2015 Nov 24;54(46):6909-16. doi: 10.1021/acs.biochem.5b01029. Epub 2015 Nov 11. Biochemistry. 2015. PMID: 26522442 Free PMC article. - Functional redundancy of steroid C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analyses.
Johnston JB, Ouellet H, Ortiz de Montellano PR. Johnston JB, et al. J Biol Chem. 2010 Nov 19;285(47):36352-60. doi: 10.1074/jbc.M110.161117. Epub 2010 Sep 15. J Biol Chem. 2010. PMID: 20843794 Free PMC article. - Cholesterol ester oxidation by mycobacterial cytochrome P450.
Frank DJ, Madrona Y, Ortiz de Montellano PR. Frank DJ, et al. J Biol Chem. 2014 Oct 31;289(44):30417-30425. doi: 10.1074/jbc.M114.602771. Epub 2014 Sep 10. J Biol Chem. 2014. PMID: 25210044 Free PMC article. - Potential drug targets in the Mycobacterium tuberculosis cytochrome P450 system.
Ortiz de Montellano PR. Ortiz de Montellano PR. J Inorg Biochem. 2018 Mar;180:235-245. doi: 10.1016/j.jinorgbio.2018.01.010. Epub 2018 Jan 12. J Inorg Biochem. 2018. PMID: 29352597 Free PMC article. Review. - The preponderance of P450s in the Mycobacterium tuberculosis genome.
McLean KJ, Clift D, Lewis DG, Sabri M, Balding PR, Sutcliffe MJ, Leys D, Munro AW. McLean KJ, et al. Trends Microbiol. 2006 May;14(5):220-8. doi: 10.1016/j.tim.2006.03.002. Epub 2006 Apr 3. Trends Microbiol. 2006. PMID: 16581251 Review.
Cited by
- Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Katariya MM, Snee M, Tunnicliffe RB, Kavanagh ME, Boshoff HIM, Amadi CN, Levy CW, Munro AW, Abell C, Leys D, Coyne AG, McLean KJ. Katariya MM, et al. Chemistry. 2023 May 22;29(29):e202203868. doi: 10.1002/chem.202203868. Epub 2023 Apr 12. Chemistry. 2023. PMID: 36912255 Free PMC article. - Microbial Steroid Production Technologies: Current Trends and Prospects.
Donova M. Donova M. Microorganisms. 2021 Dec 28;10(1):53. doi: 10.3390/microorganisms10010053. Microorganisms. 2021. PMID: 35056503 Free PMC article. - Heme and I.
Ortiz de Montellano PR. Ortiz de Montellano PR. J Biol Chem. 2015 Sep 4;290(36):21833-44. doi: 10.1074/jbc.X115.680066. Epub 2015 Jul 20. J Biol Chem. 2015. PMID: 26195628 Free PMC article. No abstract available. - Roles of cysteine in the structure and metabolic function of Mycobacterium tuberculosis CYP142A1.
Lu Y, Sun L, Pang J, Li C, Wang X, Hu X, Li G, Li X, Zhang Y, Wang H, Yang X, You X. Lu Y, et al. RSC Adv. 2022 Aug 30;12(38):24447-24455. doi: 10.1039/d2ra04257f. eCollection 2022 Aug 30. RSC Adv. 2022. PMID: 36128375 Free PMC article. - Comprehensive analysis of protein acetyltransferases of human pathogen Mycobacterium tuberculosis.
Xie L, Yang W, Fan X, Xie J. Xie L, et al. Biosci Rep. 2019 Dec 20;39(12):BSR20191661. doi: 10.1042/BSR20191661. Biosci Rep. 2019. PMID: 31820790 Free PMC article.
References
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI095437/AI/NIAID NIH HHS/United States
- GM078553/GM/NIGMS NIH HHS/United States
- R01 GM078553/GM/NIGMS NIH HHS/United States
- R01 AI074824/AI/NIAID NIH HHS/United States
- AI074824/AI/NIAID NIH HHS/United States
- R01 AI095437/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials