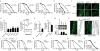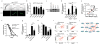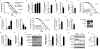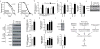Mitonuclear protein imbalance as a conserved longevity mechanism - PubMed (original) (raw)
Mitonuclear protein imbalance as a conserved longevity mechanism
Riekelt H Houtkooper et al. Nature. 2013.
Abstract
Longevity is regulated by a network of closely linked metabolic systems. We used a combination of mouse population genetics and RNA interference in Caenorhabditis elegans to identify mitochondrial ribosomal protein S5 (Mrps5) and other mitochondrial ribosomal proteins as metabolic and longevity regulators. MRP knockdown triggers mitonuclear protein imbalance, reducing mitochondrial respiration and activating the mitochondrial unfolded protein response. Specific antibiotics targeting mitochondrial translation and ethidium bromide (which impairs mitochondrial DNA transcription) pharmacologically mimic mrp knockdown and extend worm lifespan by inducing mitonuclear protein imbalance, a stoichiometric imbalance between nuclear and mitochondrially encoded proteins. This mechanism was also conserved in mammalian cells. In addition, resveratrol and rapamycin, longevity compounds acting on different molecular targets, similarly induced mitonuclear protein imbalance, the mitochondrial unfolded protein response and lifespan extension in C. elegans. Collectively these data demonstrate that MRPs represent an evolutionarily conserved protein family that ties the mitochondrial ribosome and mitonuclear protein imbalance to the mitochondrial unfolded protein response, an overarching longevity pathway across many species.
Figures
Figure 1. Lifespan regulation in BXD recombinant inbred mice
a, Life span in different BXD strains. b, Interval mapping using the BXD lifespan data reveals a strong QTL on chromosome 2, between 124–129 Mb. The red line depicts cutoff for statistical significance (p genome-wide <0.05), while the grey line represents the limit for suggestive QTLs. See also Supplementary Table 1. c, Pearson’s r correlation coefficient with corresponding p values for the covariation between BXD lifespan (x-axis) and mRNA expression of the indicated gene in the BXD eye microarrays (y-axis). Expression of Slc12a1, Mrps5 and Ttl robustly correlates with longevity (p<0.01).
Figure 2. Validation of Mrps5 as a candidate longevity gene
a, Knockdown during entire life of mrps-5, nkcc-1 or ttll-9 in C. elegans increased lifespan by 60%, 23% or 3%, respectively. See also Supplementary Table 2. b, One-way hierarchical clustering showing expression differences in gastrocnemius between young (5mo), old (25mo) and caloric restricted C57BL/6J mice. Expression of mouse _Mrp_’s decreases upon aging, and reverts with CR, whereas Slc12a1 and Ttl do not change. c, Pearson’s r correlation coefficient with corresponding p values for covariation between BXD lifespan (x-axis) and mRNA expression of eleven other _Mrp_’s (y-axis) indicates robust correlation. Principle component analysis (PCA) reveals a highly significant correlation between the Mrp gene family and BXD lifespan. d, Mrps5 strongly correlates with genes involved in OXPHOS. Red lines indicate a positive Pearson correlation coefficient of 0.7–1.0, and blue lines of 0.5–0.7.
Figure 3. mrps-5 RNAi prevents aging-associated functional decline and alters mitochondrial function
a, Knockdown of mrpl-1, mrpl-2 or mrpl-37 increased lifespan by 57%, 54%, or 41%, respectively. b, When RNAi of mrps-5 was performed during the larval stages only, lifespan increased by 48%, while RNAi started from the L4 stage had no effect. _p_≤0.001 is for larval-only versus either vector control or adult-only. c, mrps-5 or cco-1 RNAi prevented age-related changes in muscle morphology as evidenced by a pmyo-3MYO-3::GFP reporter worm highlighting myosin heavy chain. d, mrp RNAi in C. elegans decreased respiration. Respiration/worm is shown here, but respiration was similarly decreased when corrected for protein. FCCP was added at the indicated time. Values are mean±SEM (n=10), *** _p_≤0.001. e–g, mrps-5 RNAi decreased ATP levels (e, n=3), citrate synthase activity (f, n=3), and altered the ratio between nDNA (ATP5A) versus mtDNA-encoded (MTCO1) OXPHOS proteins, similar to cco-1, but not mev-1 (g, n=4). * _p_≤0.05. h, mrps-5 RNAi resulted in fragmented mitochondria, as visualized using the pmyo-3::mito::GFP reporter, which expresses mitochondria-targeted GFP driven by the muscle-specific myo-3 promoter. i, mrps-5 RNAi increased mean lifespan by 40%. j-m, mrps-5 RNAi extends lifespan of daf-16(mu86) (j), sir-2.1(ok434) (k), aak-2(ok524) (l), mev-1(kn1) (m) mutants by 37%, 40%, 69%, 112%. n, Knockdown of cco-1 does not extend lifespan of mrps-5 RNAi worms. See Supplementary Table 2 and Fig. 4.
Figure 4. mrp genes confer longevity effects through UPRmt
a, RNAi of mrp genes induced UPRmt (hsp-6::GFP reporter), similar to cco-1 knockdown. Worms are synchronized at day 1 of adulthood. b, Quantification of UPRmt upon knockdown of mrp or cco-1. (n=4) c, mrps-5 and cco-1, but not mev-1, RNAi, induce UPRmt as reflected by the induction of HSP-6-GFP protein. d, Combined RNAi of mrps-5 and mev-1 synergistically increased UPRmt, while combined cco-1/mrps-5 RNAi did not further increase UPRmt (n=6). e, Knockdown of different mrp genes results in different levels of UPRmt, which correlates with mean lifespan (n=33–61 worms for lifespan, n=3 for GFP). f–h, Epistasis with UPRmt regulator haf-1. Double RNAi of mrps-5 and haf-1 partially prevented lifespan extension (f), UPRmt (g, n=5), and reduction in respiration (h, n=10), compared to mrps-5 RNAi alone. i, In various tissues of mouse crosses, Ubl5, Abcb10 and Hspd1 expression correlated with Mrp expression. j, Hspd1 (HSP60) ties in a correlation network with Mrp and OXPHOS genes. Connecting lines indicate a Pearson correlation coefficient of 0.75–1.0. Bar graphs are mean±SEM, _* p_≤0.05; ** _p_≤0.01; *** _p_≤0.001. See also Supplementary Fig. 5–7 and Table 3.
Figure 5. Specific antibiotics extend lifespan by phenocopying mrps-5 knockdown
a, Effects on worm lifespan of doxycycline (30μg/mL), compared to carbenicilin (30μg/mL) or vehicle. _p_≤0.001 refers to statistical significance of doxycycline compared to either vehicle or the carbenicilin control. Antibiotics were given throughout life in panels a–e. b, The effects of doxycycline on lifespan are dose-dependent. c–e, Doxycycline induced UPRmt (c, n=5) and reduced respiration (d, n=10), without changing citrate synthase activity (e, n=3). f–k, When treated only during larval development, doxycycline (6μg/mL; f–h) and chloramphenicol (100μg/mL; i–k) extend lifespan (f, i), induced UPRmt (g, j, n=5) and reduced respiration (h, k, n=6). l, Doxycycline alters the ratio between nDNA- (ATP5A) and mtDNA-encoded (MTCO1) OXPHOS proteins in worms. m, Doxycycline decreased respiration in a cultured hepatocyte cell line (n=5), n, induced Hsp60 transcription, as measured using an Hsp60 promoter reporter (n=8), o, increased HSP60 protein expression and altered the ratio of nDNA- (UQCRC2) versus mtDNA- (MTCOI) encoded proteins (n=2). p, Doxycycline increased HSP60 protein and altered the ratio of nDNA- versus mtDNA-encoded proteins in primary murine hepatocytes. q, Doxycycline (50 mpkd) for 10 days in C57BL/6N mice decreased oxygen consumption (n=10). See also Supplementary Table 2.
Figure 6. Conserved function of mitonuclear protein imbalance and UPRmt in longevity
a, Rapamycin (1nM) extends worm lifespan in a _ubl-5_-dependent manner, and b, _ubl-5_-dependently induced UPRmt (hsp-6::GFP) but not UPRER (hsp-4::GFP) (n=4). c–e, Rapamycin increased respiration (c, n=10) and ATP content but not citrate synthase activity (d, n=3) and induced mitonuclear protein imbalance (e). f–h, In mouse hepatocytes, rapamycin induces mitonuclear protein imbalance (f–g) and induces UPRmt as shown at the protein (f–g, n=3), and transcriptional (h, n=8) level. i, Resveratrol (25 μM) induced mitonuclear protein imbalance in mouse hepatocytes (n=4). j, Hypothetical scheme of the mechanism by which reduced Mrp expression (during aging or genetic inactivation), specific antibiotics, ethidium bromide and rapamycin and resveratrol extend lifespan by inducing UPRmt. Bar graphs are expressed as mean±SEM, _* p_≤0.05; *** _p_≤0.001. See also Supplementary Table 2.
Comment in
- Ageing: beneficial miscommunication.
Wolff S, Dillin A. Wolff S, et al. Nature. 2013 May 23;497(7450):442-3. doi: 10.1038/497442a. Nature. 2013. PMID: 23698438 No abstract available. - Ageing: an ageing balancing act.
Stower H. Stower H. Nat Rev Genet. 2013 Jul;14(7):442. doi: 10.1038/nrg3517. Epub 2013 Jun 4. Nat Rev Genet. 2013. PMID: 23732334 No abstract available.
Similar articles
- Fragile lifespan expansion by dietary mitohormesis in C. elegans.
Tauffenberger A, Vaccaro A, Parker JA. Tauffenberger A, et al. Aging (Albany NY). 2016 Jan;8(1):50-61. doi: 10.18632/aging.100863. Aging (Albany NY). 2016. PMID: 26764305 Free PMC article. - A Conserved Mito-Cytosolic Translational Balance Links Two Longevity Pathways.
Molenaars M, Janssens GE, Williams EG, Jongejan A, Lan J, Rabot S, Joly F, Moerland PD, Schomakers BV, Lezzerini M, Liu YJ, McCormick MA, Kennedy BK, van Weeghel M, van Kampen AHC, Aebersold R, MacInnes AW, Houtkooper RH. Molenaars M, et al. Cell Metab. 2020 Mar 3;31(3):549-563.e7. doi: 10.1016/j.cmet.2020.01.011. Epub 2020 Feb 20. Cell Metab. 2020. PMID: 32084377 Free PMC article. - The mitochondrial unfolded protein response and increased longevity: cause, consequence, or correlation?
Bennett CF, Kaeberlein M. Bennett CF, et al. Exp Gerontol. 2014 Aug;56:142-6. doi: 10.1016/j.exger.2014.02.002. Epub 2014 Feb 8. Exp Gerontol. 2014. PMID: 24518875 Free PMC article. Review. - Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans.
Bennett CF, Vander Wende H, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M. Bennett CF, et al. Nat Commun. 2014 Mar 24;5:3483. doi: 10.1038/ncomms4483. Nat Commun. 2014. PMID: 24662282 Free PMC article. - Mitochondrial dysfunction, aging, and the mitochondrial unfolded protein response in Caenorhabditis elegans.
Haynes CM, Hekimi S. Haynes CM, et al. Genetics. 2022 Nov 30;222(4):iyac160. doi: 10.1093/genetics/iyac160. Genetics. 2022. PMID: 36342845 Free PMC article. Review.
Cited by
- Single cell analysis revealed that two distinct, unique CD4+ T cell subsets were increased in the small intestinal intraepithelial lymphocytes of aged mice.
Yonemoto Y, Nemoto Y, Morikawa R, Shibayama N, Oshima S, Nagaishi T, Mizutani T, Ito G, Fujii S, Okamoto R. Yonemoto Y, et al. Front Immunol. 2024 Jan 22;15:1340048. doi: 10.3389/fimmu.2024.1340048. eCollection 2024. Front Immunol. 2024. PMID: 38327516 Free PMC article. - Emerging therapeutic roles for NAD(+) metabolism in mitochondrial and age-related disorders.
Srivastava S. Srivastava S. Clin Transl Med. 2016 Dec;5(1):25. doi: 10.1186/s40169-016-0104-7. Epub 2016 Jul 27. Clin Transl Med. 2016. PMID: 27465020 Free PMC article. Review. - The diabetic microenvironment causes mitochondrial oxidative stress in glomerular endothelial cells and pathological crosstalk with podocytes.
Casalena GA, Yu L, Gil R, Rodriguez S, Sosa S, Janssen W, Azeloglu EU, Leventhal JS, Daehn IS. Casalena GA, et al. Cell Commun Signal. 2020 Jul 8;18(1):105. doi: 10.1186/s12964-020-00605-x. Cell Commun Signal. 2020. PMID: 32641054 Free PMC article. - The Convergence of Systems and Reductionist Approaches in Complex Trait Analysis.
Williams EG, Auwerx J. Williams EG, et al. Cell. 2015 Jul 2;162(1):23-32. doi: 10.1016/j.cell.2015.06.024. Cell. 2015. PMID: 26140590 Free PMC article. Review. - N1-methylation of adenosine (m1A) in ND5 mRNA leads to complex I dysfunction in Alzheimer's disease.
Jörg M, Plehn JE, Kristen M, Lander M, Walz L, Lietz C, Wijns J, Pichot F, Rojas-Charry L, Wirtz Martin KM, Ruffini N, Kreim N, Gerber S, Motorin Y, Endres K, Rossmanith W, Methner A, Helm M, Friedland K. Jörg M, et al. Mol Psychiatry. 2024 May;29(5):1427-1439. doi: 10.1038/s41380-024-02421-y. Epub 2024 Jan 29. Mol Psychiatry. 2024. PMID: 38287100 Free PMC article.
References
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. - PubMed
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20-DA 21131/DA/NIDA NIH HHS/United States
- UO1AA13499/AA/NIAAA NIH HHS/United States
- R01 AG043930/AG/NIA NIH HHS/United States
- P20 DA021131/DA/NIDA NIH HHS/United States
- 231138/ERC_/European Research Council/International
- U01 AA013499/AA/NIAAA NIH HHS/United States
- U01AA14425/AA/NIAAA NIH HHS/United States
- U01 AA014425/AA/NIAAA NIH HHS/United States
- R01AG043930/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases