MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer - PubMed (original) (raw)
. 2014 Apr 14;25(4):469-83.
doi: 10.1016/j.ccr.2014.03.006.
Chiara Braconi 2, Pierluigi Gasparini 3, Claudio Murgia 4, Andrea Lampis 2, Viola Paulus-Hock 2, Jonathan R Hart 5, Lynn Ueno 5, Sergei I Grivennikov 6, Francesca Lovat 3, Alessio Paone 3, Luciano Cascione 3, Khlea M Sumani 3, Angelo Veronese 7, Muller Fabbri 3, Stefania Carasi 3, Hansjuerg Alder 3, Giovanni Lanza 8, Roberta Gafa' 8, Mary P Moyer 9, Rachel A Ridgway 4, Julia Cordero 4, Gerard J Nuovo 3, Wendy L Frankel 10, Massimo Rugge 11, Matteo Fassan 11, Joanna Groden 3, Peter K Vogt 5, Michael Karin 6, Owen J Sansom 4, Carlo M Croce 12
Affiliations
- PMID: 24735923
- PMCID: PMC3995091
- DOI: 10.1016/j.ccr.2014.03.006
MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer
Nicola Valeri et al. Cancer Cell. 2014.
Abstract
MicroRNA deregulation is frequent in human colorectal cancers (CRCs), but little is known as to whether it represents a bystander event or actually drives tumor progression in vivo. We show that miR-135b overexpression is triggered in mice and humans by APC loss, PTEN/PI3K pathway deregulation, and SRC overexpression and promotes tumor transformation and progression. We show that miR-135b upregulation is common in sporadic and inflammatory bowel disease-associated human CRCs and correlates with tumor stage and poor clinical outcome. Inhibition of miR-135b in CRC mouse models reduces tumor growth by controlling genes involved in proliferation, invasion, and apoptosis. We identify miR-135b as a key downsteam effector of oncogenic pathways and a potential target for CRC treatment.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
Figure 1
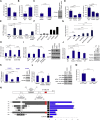
MiR-135b Upregulation in Mouse and Human CRC (A) Genome-wide miR expression analysis in tumor and normal adjacent colon tissues from CPC;Apc and AOM/DSS treated mice. Selected deregulated miRNAs in cancer relative to normal tissues are plotted against the p value. (B and C) MiR-135b expression by RT-PCR (B) and ISH (C) in both CRC mouse models. Scale bars, 200 um (100 um in the magnification). (D) MiR-135b expression was assessed in human tissues. Paired analysis (a) and miR expression according to stage (b) in sporadic CRC (n = 62) are shown. Reads for miR-135b from the TCGA miR-seq data set (n = 392) are shown (c). miR-135b upregulation in three different cohorts of IBD-associated CRC (n = 31): OSU (n = 9; d), University of Ferrara (n = 9; e), and University of Padua (n = 13; f). (E) ISH for miR-135b in human cancer and normal epithelial cells. Scale bars, 200 um (50 um in the magnification). (F) Prognosis of sporadic CRC patients according to miR-135b expression: (a) overall survival (OS) in the entire cohort, (b) relapse-free survival (RFS) in stage I-II-III CRC, (c) RFS in stage II-III only CRC and (d) in stage II only CRC. High or low miR-135b expression was defined as low if Log2-miR-135b-ratio (cancer/normal) ≤2 or high if Log2-miR-135b-ratio > 2. See also Tables S1–S3 and Figure S1.
Figure 2
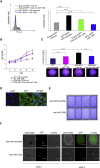
MiR-135b Overexpression Is Associated with Mutations in Specific CRC Pathways (A) miR-135b expression following transfection with a plasmid encoding the APC CDS in APC mutant SW480 cells. (B) miR-135b expression following APC silencing in normal colon (NCM 460) epithelial cells. (C) miR-135b expression (left) following overexpression of CTNNB1 (right) in NCM 460 normal epithelial cells. (D) miR-135b expression following silencing of TCF4 and LEF1 transcription in APC mutant SW480 cells (left) or CTNNB1 mutant HCT-116 cells (right). (E–G) miR-135b expression in MEFs (E), human normal and CRC cell lines (F), and tumor-derived organoids from GEMMs (G) harboring different rounds of mutations in genes commonly mutated in human CRC. (H–J) miR-135b expression in PI3KCA WT and mutant HCT-116 cells after serum starvation (H), treatment with LY-294002 (I), and transfection with specific siRNAs to FoxO1 and FoxO3A (J). (K) miR-135b expression in parental SW480 and SW620 CRC cell lines after treatment with dasatinib. (L) miR-135b expression in Src-MEFs after treatment with PI3K or MEK1/2 inhibitors. (M) miR-135b expression in tumor derived from AhCre Apc fl/fl Src +/+ and AhCre Apc fl/fl Src fl/fl mice. (N) Luciferase reporter assay was performed after LiCl treatment using different vectors containing predicted TCF4 binding sites upstream of the pre–miR-135b; −1 position corresponds to the 5′ terminus of the miR-135b hairpin. Putative TCF4 responsive sequences (CTTTGTT)) are indicated in the gray boxes, and the miR-135b sequence is in red. Deletion of the TCF4 binding site is represented by a X. Human or mouse miR-135b expression was assessed by RT-PCR and normalized to that of RNU48 or SNU234, respectively. Bars represent the mean and SD of three experiments; p values are reported within the figures. See also Figure S2.
Figure 3
MiR-135b Mediates the Cancer Phenotype Induced by APC and PI3KCA Mutations (A) APC mutant SW480 cells were transfected with pre-miR-135b, LNA anti-miR-135b, or controls in combination with a plasmid encoding the APC CDS or an empty vector. Apoptotic cells were measured by flow cytometer (left) and quantitated (right). (B) Isogenic PI3KCA WT and mutant HCT-116 cells were transfected with pre-miR-135b, LNA anti-miR-135b, or relative controls. Cell viability was measured at selected time points. (C) Isogenic PI3KCA mutant and WT HCT-116 cells overexpressing miR-135b or anti-miR-135b were plated in soft agar and grown in low fetal bovine serum conditions. Colonies greater than 2 mm in size were counted and quantitated. Representative images are shown. (D–F) v-SRC transformed MEFs were infected with lentiviruses encoding an anti-miR-135b or a scramble hairpin. Cells were selected with puromycin and checked for viral integration using GFP (scale bars, 20 um; D). Cells were grown in soft agar and colonies were counted after 4 weeks. On the far left of the plate, no cells were seeded as negative control (E). GFP was used to monitor viral integration during for the entire duration of the experiment (F). Scale bars, 200 um (week 1) and 400 um (week 4). Bars represent the mean and SD of three experiments; p values are reported within the figures. See also Tables S4 and S5 and Figure S3.
Figure 4
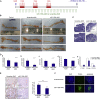
miR-135b Inhibition Rescues Apc-Induced Phenotype In Vivo (A) Conditional Apc deletion induced by b-napthoflavone injection in AhCre + Apc +/+ mice on days 1 and 3. miR-135b-AMO or scrambled-AMO were injected on days 2 and 4. Mice (n = 6 for each group) were euthanized on day 4. (B) Graphs show total cells per crypt (left) and proliferation index (ratio between BrDU-positive cells and total cells per crypt; right) in AhCre + Apc +/+ and AhCre + Apc fl/fl mice treated with scramble-AMO or anti-135b-AMO. BrDU was injected in three mice per group 2 hr prior to euthanization. Error bars represent SD; p values are shown within the graphs. Magnification bars, 100 um (top), 50 um (bottom). (C) Hematoxylin and eosin (H&E) and BrDU-stained sections for AhCre+Apc+/+ and AhCre+Apcfl/fl mice treated with scramble-AMO or anti-135b-AMO. Bars show size of the crypts. Magnification bars, 100 um (top), 50 um (bottom). See also Figure S4.
Figure 5
Anti-miR-135b Exerts an Antitumor Effect In Vivo in the AOM/DSS Model (A–C) Overview of the study (A): AOM was given once, followed by periodic administration of DSS in water. MiR-135b-AMO or scrambled-AMO were given twice a week for 100 days. Mice were treated with miR-135b-AMO (n = 8), scrambled-AMO (n = 8), or left untreated (n = 8). Following euthanization, macroscopic (B) and microscopic (C) analysis of the tumors was performed. Scale bars, 200 um (top), 100 um (bottom). (D) Statistical analysis of tumor number (multiplicity, a), tumor number for tumors >2 mm (b), tumor volume (load, c), average size (d), and tumor size distribution in the two groups (e). (E and F) Cancer tissues from mice treated with miR-135b-AMO or scrambled-AMO were analyzed. (E) Ki-67 expression was assessed with immunohistochemistry (a) and quantitated (b; scale bars, 200 um [top], 100 um [bottom]). (F) Apoptosis was assessed by immunofluorescence (scale bar, 200 um). See also Figure S5.
Figure 6
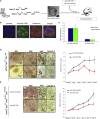
miR-135b Inhibition Causes Tumor Inhibition in Organoids Derived from CRC GEMMs (A and B) Tumor-derived organoids from Apc fl/fl /Pten fl/fl and Apc fl/fl /Kras G12/+ /P53 R172H/fl (A) infected with lentiviruses encoding anti-miR-135b or scramble hairpins (B) were xenotransplanted in nude mice. Tumors were measured twice a week for 8 weeks or until ulceration. (C) miR-135b expression was assessed by RT-PCR in tumor-derived organoids after infection. Bars represent the mean and SD of three experiments; p < 0.01. (D) Microscopic analysis (left) and tumor growth (right) in Apc fl/fl /Pten fl/fl organoids. (E) Microscopic analysis (left) and tumor growth (right) in Apc fl/fl /Kras G12/+ /P53 R172H/fl organoids. Results are expressed as the mean percentage of change in tumor volume for each group of mice, ± SEM. See also Figure S6 and Tables S6 and S7.
Figure 7
MiR-135b Controls Tumor-Suppressor Genes (A) Analysis of a cancer gene-associated panel in 460NCM cell transfected with pre-miR-135b or scrambled probes. (B) APC (b), TGFβR2 (c), and DAPK1 (d) mRNA expression analyzed by RT-PCR following miR-135b overexpression (a). (C) In silico target prediction for miR-135b binding sites in APC, TGFβR2, and DAPK1 3′UTRs. (D) Luciferase experiments for TGFβR2, DAPK1, and APC in NCM460 cells transfected with target-gene-Luc-WT or target-gene-Luc mutant, pre-miR-135b, or scrambled miR. (E) DAPK1 protein expression by WB following transfection with pre-miR-135b and specific siRNA. (F) DAPK1 mRNA expression in dysplasia and cancer compared to normal adjacent tissues in IBD-associated CRC. (G and H) TGFβR2 and p21 protein expression assessed following miR-135b overexpression and TGF-β stimulation (G). Apoptosis measured by caspase 3/7 assay following TGF-β stimulation (H). (I) TGFβR2 mRNA expression in dysplasia and cancer compared to normal adjacent tissues in IBD-associated CRC. (J) IL8 mRNA expression following miR-135b overexpression in NCM460 cells. (K) Luciferase assay in NCM460 transfected with FIH-Luc-WT or FIH-gene-Luc mutant vectors, pre-miR-135b, or scrambled miR. MiR-135b seed region in FIH 3′UTR is shown. (L and M) FIH mRNA (L) and protein (M) expression following miR-135b overexpression in 460NCM cells. (N) FIH, IL8, and VEGFA mRNA following siRNA transfection. ∗p < 0.05. (O) Tube formation in HUVEC cells cultured with media from scrambled or miR-135b transfected cells: representative picture (left) and normalization (right) are shown. Bars represent the mean and SD of three experiments; p values are reported within the figures. See also Table S8 and Figure S7.
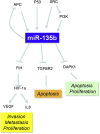
Figure 8
MiR-135b Is a Key Oncogenic Hub Mediating the Cancer Phenotype Downstream of Genes Frequently Mutated in CRC Schematic representation of genetic aberrations promoting miR-135b overexpression and miR-135b downstream targets.
Similar articles
- Upregulation of microRNA-135b and microRNA-182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway.
Liu B, Liu Y, Zhao L, Pan Y, Shan Y, Li Y, Jia L. Liu B, et al. Mol Carcinog. 2017 Dec;56(12):2669-2680. doi: 10.1002/mc.22710. Epub 2017 Aug 21. Mol Carcinog. 2017. PMID: 28767179 - MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1.
Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM, Yang PC. Lin CW, et al. Nat Commun. 2013;4:1877. doi: 10.1038/ncomms2876. Nat Commun. 2013. PMID: 23695671 - Knockdown of Mir-135b Sensitizes Colorectal Cancer Cells to Oxaliplatin-Induced Apoptosis Through Increase of FOXO1.
Qin Y, Li L, Wang F, Zhou X, Liu Y, Yin Y, Qi X. Qin Y, et al. Cell Physiol Biochem. 2018;48(4):1628-1637. doi: 10.1159/000492284. Epub 2018 Aug 2. Cell Physiol Biochem. 2018. PMID: 30071508 - Differential expression of microRNAs in tumors from chronically inflamed or genetic (APC(Min/+)) models of colon cancer.
Necela BM, Carr JM, Asmann YW, Thompson EA. Necela BM, et al. PLoS One. 2011 Apr 12;6(4):e18501. doi: 10.1371/journal.pone.0018501. PLoS One. 2011. PMID: 21532750 Free PMC article. - MicroRNA-135b mainly functions as an oncogene during tumor progression.
Tolue Ghasaban F, Taghehchian N, Zangouei AS, Keivany MR, Moghbeli M. Tolue Ghasaban F, et al. Pathol Res Pract. 2024 Oct;262:155547. doi: 10.1016/j.prp.2024.155547. Epub 2024 Aug 15. Pathol Res Pract. 2024. PMID: 39151250 Review.
Cited by
- Three-layer heterogeneous network based on the integration of CircRNA information for MiRNA-disease association prediction.
Qu J, Liu S, Li H, Zhou J, Bian Z, Song Z, Jiang Z. Qu J, et al. PeerJ Comput Sci. 2024 Jun 10;10:e2070. doi: 10.7717/peerj-cs.2070. eCollection 2024. PeerJ Comput Sci. 2024. PMID: 38983241 Free PMC article. - MicroRNAs and Apoptosis in Colorectal Cancer.
Wang H. Wang H. Int J Mol Sci. 2020 Jul 28;21(15):5353. doi: 10.3390/ijms21155353. Int J Mol Sci. 2020. PMID: 32731413 Free PMC article. Review. - Magnetic Resonance Imaging for Translational Research in Oncology.
Fiordelisi MF, Cavaliere C, Auletta L, Basso L, Salvatore M. Fiordelisi MF, et al. J Clin Med. 2019 Nov 6;8(11):1883. doi: 10.3390/jcm8111883. J Clin Med. 2019. PMID: 31698697 Free PMC article. Review. - Cancer-associated fibroblasts derived extracellular vesicles promote angiogenesis of colorectal adenocarcinoma cells through miR-135b-5p/FOXO1 axis.
Dai X, Xie Y, Dong M. Dai X, et al. Cancer Biol Ther. 2022 Dec 31;23(1):76-88. doi: 10.1080/15384047.2021.2017222. Cancer Biol Ther. 2022. PMID: 35100092 Free PMC article. - Development of miRNA-based therapeutic approaches for cancer patients.
Takahashi RU, Prieto-Vila M, Kohama I, Ochiya T. Takahashi RU, et al. Cancer Sci. 2019 Apr;110(4):1140-1147. doi: 10.1111/cas.13965. Epub 2019 Feb 26. Cancer Sci. 2019. PMID: 30729639 Free PMC article. Review.
References
- Aust D.E., Terdiman J.P., Willenbucher R.F., Chang C.G., Molinaro-Clark A., Baretton G.B., Loehrs U., Waldman F.M. The APC/beta-catenin pathway in ulcerative colitis-related colorectal carcinomas: a mutational analysis. Cancer. 2002;94:1421–1427. - PubMed
- Biswas S., Chytil A., Washington K., Romero-Gallo J., Gorska A.E., Wirth P.S., Gautam S., Moses H.L., Grady W.M. Transforming growth factor beta receptor type II inactivation promotes the establishment and progression of colon cancer. Cancer Res. 2004;64:4687–4692. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 CA152758/CA/NCI NIH HHS/United States
- R00 DK088589/DK/NIDDK NIH HHS/United States
- 18052/CRUK_/Cancer Research UK/United Kingdom
- P01 DK035108/DK/NIDDK NIH HHS/United States
- K99 DK088589/DK/NIDDK NIH HHS/United States
- DK035108/DK/NIDDK NIH HHS/United States
- P30 CA016058/CA/NCI NIH HHS/United States
- R01 AI043477/AI/NIAID NIH HHS/United States
- CSO_/Chief Scientist Office/United Kingdom
- 1R01 CA078230/CA/NCI NIH HHS/United States
- AI043477/AI/NIAID NIH HHS/United States
- RC2 CA148302/CA/NCI NIH HHS/United States
- K99-DK088589/DK/NIDDK NIH HHS/United States
- 12481/CRUK_/Cancer Research UK/United Kingdom
- R01 CA078230/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous