The natural chemopreventive agent sulforaphane inhibits STAT5 activity - PubMed (original) (raw)
The natural chemopreventive agent sulforaphane inhibits STAT5 activity
Sophia Pinz et al. PLoS One. 2014.
Abstract
Signal transducer and activator of transcription STAT5 is an essential mediator of cytokine, growth factor and hormone signaling. While its activity is tightly regulated in normal cells, its constitutive activation directly contributes to oncogenesis and is associated to a number of hematological and solid tumor cancers. We previously showed that deacetylase inhibitors can inhibit STAT5 transcriptional activity. We now investigated whether the dietary chemopreventive agent sulforaphane, known for its activity as deacetylase inhibitor, might also inhibit STAT5 activity and thus could act as a chemopreventive agent in STAT5-associated cancers. We describe here sulforaphane (SFN) as a novel STAT5 inhibitor. We showed that SFN, like the deacetylase inhibitor trichostatin A (TSA), can inhibit expression of STAT5 target genes in the B cell line Ba/F3, as well as in its transformed counterpart Ba/F3-1*6 and in the human leukemic cell line K562 both of which express a constitutively active form of STAT5. Similarly to TSA, SFN does not alter STAT5 initial activation by phosphorylation or binding to the promoter of specific target genes, in favor of a downstream transcriptional inhibitory effect. Chromatin immunoprecipitation assays revealed that, in contrast to TSA however, SFN only partially impaired the recruitment of RNA polymerase II at STAT5 target genes and did not alter histone H3 and H4 acetylation, suggesting an inhibitory mechanism distinct from that of TSA. Altogether, our data revealed that the natural compound sulforaphane can inhibit STAT5 downstream activity, and as such represents an attractive cancer chemoprotective agent targeting the STAT5 signaling pathway.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
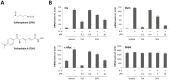
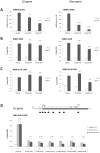
Figure 1. Sulforaphane (SFN) treatment inhibits IL-3-mediated induction of STAT5 target genes in Ba/F3 cells in a dose-dependent manner.
(A) Structure of the natural compound sulforaphane (SFN) and of the synthetic deacetylase inhibitor trichostatin A (TSA) used in this study. (B) Ba/F3 cells were pre-treated 30 minutes with DMSO (vehicle), 0.2 µM TSA, 0.4, 2 or 10 µM SFN and further stimulated 30 minutes with 5 ng/mL IL-3. Following cell harvest, expression of the STAT5 target genes Cis, Osm, c-Myc and of the housekeeping gene 36b4 were measured by quantitative RT-PCR, as described in Materials and Methods. Similarly to TSA, SFN inhibits IL-3-mediated induction of STAT5-regulated genes.
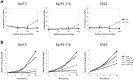
Figure 2. Effect of SFN treatment on cytotoxicity and viability of normal (Ba/F3) and transformed (Ba/F3-1*6, K562) cells.
(A) The WST-1 reagent was added to cells following 30 minutes of pre-treatment with 0.001, 0.01, 0.1 and 1 µM TSA or with 0.1, 1, 10 and 100 µM SFN. IL-3 (5 ng/mL) was supplemented to rested Ba/F3 cells at the same time as the WST-1 reagent to mimic the IL-3 stimulation conditions used in other assays. OD measurement was performed after 90 minutes incubation with the WST-1 reagent, and the percentage of cytotoxicity was normalized to the vehicle control. (B) Growing Ba/F3, Ba/F3-1*6 and K562 cells were incubated for 24 and 48 hours in the presence of the indicated concentrations of TSA and SFN. Cell viability was measured by Trypan Blue exclusion assay.
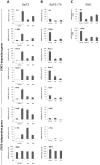
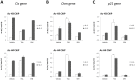
Figure 3. SFN treatment inhibits STAT5 constitutive activity in the transformed cell lines Ba/F3-1*6 and K562.
Ba/F3 (A), its transformed counterpart Ba/F3-1*6 (B) and human leukemic K562 (C) cells were treated 90 minutes with DMSO (vehicle), 0.2 µM TSA, 10 µM SFN or 1 µM Imatinib. Ba/F3 cells (A) were stimulated with 5 ng/mL IL-3 for 60 minutes following 30 minutes of drug pre-treatment. Expression of STAT5-dependent (Cis, c-Myc, Pim-1, Socs-1, Osm,) and -independent (JunB, c-Fos, 36b4) genes was analyzed by quantitative RT-PCR. Gene expression data were normalized to cDNA levels derived from mouse ribosomal S9 (A, B) or human Lamin A/C (LMNA) (C) mRNAs. (A, B) The Y-axis scales were adjusted to allow a direct comparison of relative expression levels in Ba/F3 and Ba/F3-1*6 cells.
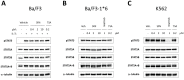
Figure 4. SFN treatment does not affect STAT5 phosphorylation.
Ba/F3 (A), Ba/F3-1*6 (B) and K562 (C) cells were treated 60 minutes with DMSO (vehicle) or the indicated concentrations of TSA, SFN or Imatinib. Ba/F3 cells (A) were stimulated with 5 ng/mL IL-3 for 30 minutes following 30 minutes of drug pre-treatment. Whole-cell Brij protein lysates were analyzed by Western blot using antibodies specific for phospho-STAT5 (pSTAT5), STAT5A, STAT5B, STAT5A and B, and α-tubulin (loading control).
Figure 5. STAT5 binding and RNA polymerase II recruitment to the promoter of STAT5 target genes are marginally affected by SFN treatment.
Ba/F3 cells were pre-treated 30 minutes with DMSO (vehicle), 10 µM or 20 µM SFN and further stimulated with 5 ng/mL IL-3 for 30 minutes. Cells were harvested for both gene expression analysis of the Cis and Osm genes by quantitative RT-PCR (A) and for chromatin immunoprecipitation (ChIP) (B–D). ChIP was performed using antibodies directed against STAT5 (B) or RNA polymerase II (RNA Pol II; C, D) proteins. Co-precipitated genomic DNA was analyzed by quantitative PCR using primers specific for the STAT5 binding sites (amplicons A and I in Figure S2) (STAT5 ChIP; B) or the transcription start site (amplicons B and J in Figure S2) (RNA Pol II ChIP; C) of the mouse Cis and Osm genes, as well as with primers spanning the open reading frame of the Cis gene (RNA Pol II ChIP; D). Schematic representation of the Cis gene with its transcribed region (dark grey arrow), the coding sequence (white arrow with exons in light grey), the four STAT5 binding sites within its proximal promoter region, and the quantitative PCR amplicons investigated (A to H-labeled black boxes) is shown in (D). The RNA polymerase II occupancy along the transcribed region of the Cis gene is slightly but consistently reduced in SFN-treated cells. Two-tailed paired Student's t-test, SFN-treated compared to vehicle control (IL-3-stimulated); *P<0.05, **P<0.005, ***P<0.001, ****P<0.0001; ns, not significant.
Figure 6. SFN treatment does not affect global histone acetylation level in Ba/F3 cells.
Ba/F3 cells were treated for the indicated times with either 0.2 µM TSA or 10 µM SFN. Whole-cell Freeze-Thaw protein lysates were analyzed by Western blot using antibodies specific for acetylated histone H3 (Ac-H3) and H4 (Ac-H4) and for total histone H3 proteins as a reference. While global histone acetylation was markedly increased in cells treated with TSA, no apparent effect was detected upon SFN treatment.
Figure 7. SFN treatment does not affect histone acetylation at the promoters of STAT5 target (Cis, Osm) and control (p21) genes.
Ba/F3 cells were pre-treated 30 minutes with DMSO (vehicle), 0.2 µM TSA or 10 µM SFN and further stimulated 30 minutes with 5 ng/mL IL-3. Chromatin immunoprecipitation (ChIP) was performed using antibodies directed against acetylated histone H3 (Ac-H3) and H4 (Ac-H4) and against histone H3 proteins (total H3). Co-precipitated genomic DNA was analyzed by quantitative PCR using primers specific for the transcription start sites of the mouse Cis (A) and Osm (B) genes (amplicons B and J respectively in Figure S2), as well as for the proximal promoter region of the mouse p21 gene (amplicon K in Figure S2) as a control (C). Ac-H3 and Ac-H4 ChIP data were normalized to total Histone H3, to more accurately estimate histone acetylation levels at the investigated gene loci. Corresponding raw ChIP data for Ac-H3, Ac-H4 and H3 immunoprecipitations (expressed as % of input DNA) are shown in Figure S6. While histone acetylation levels were dramatically affected by TSA at all three gene loci, no major change in histone H3 and H4 acetylation was monitored in SFN-treated cells.
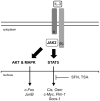
Figure 8. Model of inhibition of STAT5 activity by SFN in Ba/F3 cells.
IL-3 binding to its receptor leads to activation of the receptor-associated JAK2 tyrosine kinase. In turn, JAK2 activates the downstream STAT5, MAPK and AKT pathways via phosphorylation (broad arrows), resulting in induced transcription of downstream target genes (thin arrows). We showed that, similarly to TSA, SFN inhibits induction of STAT5 target genes without interfering with STAT5 initial activation (phosphorylation) and binding to DNA. In contrast to TSA however, SFN does not affect histone acetylation, neither globally nor locally at specific gene loci, and only moderately interferes with recruitment of the transcriptional machinery, suggesting an alternative mechanism of transcriptional inhibition, independent of deacetylase activity. JunB expression was also inhibited by SFN in Ba/F3 cells, although via a MAPK-independent mechanism.
Similar articles
- Deacetylase inhibitors repress STAT5-mediated transcription by interfering with bromodomain and extra-terminal (BET) protein function.
Pinz S, Unser S, Buob D, Fischer P, Jobst B, Rascle A. Pinz S, et al. Nucleic Acids Res. 2015 Apr 20;43(7):3524-45. doi: 10.1093/nar/gkv188. Epub 2015 Mar 13. Nucleic Acids Res. 2015. PMID: 25769527 Free PMC article. - The Chemopreventive Phytochemical Moringin Isolated from Moringa oleifera Seeds Inhibits JAK/STAT Signaling.
Michl C, Vivarelli F, Weigl J, De Nicola GR, Canistro D, Paolini M, Iori R, Rascle A. Michl C, et al. PLoS One. 2016 Jun 15;11(6):e0157430. doi: 10.1371/journal.pone.0157430. eCollection 2016. PLoS One. 2016. PMID: 27304884 Free PMC article. - Chromatin acetylation and remodeling at the Cis promoter during STAT5-induced transcription.
Rascle A, Lees E. Rascle A, et al. Nucleic Acids Res. 2003 Dec 1;31(23):6882-90. doi: 10.1093/nar/gkg907. Nucleic Acids Res. 2003. PMID: 14627821 Free PMC article. - Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: studies with sulforaphane and garlic organosulfur compounds.
Nian H, Delage B, Ho E, Dashwood RH. Nian H, et al. Environ Mol Mutagen. 2009 Apr;50(3):213-21. doi: 10.1002/em.20454. Environ Mol Mutagen. 2009. PMID: 19197985 Free PMC article. Review. - Histone deacetylases as targets for dietary cancer preventive agents: lessons learned with butyrate, diallyl disulfide, and sulforaphane.
Myzak MC, Dashwood RH. Myzak MC, et al. Curr Drug Targets. 2006 Apr;7(4):443-52. doi: 10.2174/138945006776359467. Curr Drug Targets. 2006. PMID: 16611031 Review.
Cited by
- Novel molecules as the emerging trends in cancer treatment: an update.
Sekar P, Ravitchandirane R, Khanam S, Muniraj N, Cassinadane AV. Sekar P, et al. Med Oncol. 2022 Jan 4;39(2):20. doi: 10.1007/s12032-021-01615-6. Med Oncol. 2022. PMID: 34982273 Review. - How Intrinsic Molecular Dynamics Control Intramolecular Communication in Signal Transducers and Activators of Transcription Factor STAT5.
Langenfeld F, Guarracino Y, Arock M, Trouvé A, Tchertanov L. Langenfeld F, et al. PLoS One. 2015 Dec 30;10(12):e0145142. doi: 10.1371/journal.pone.0145142. eCollection 2015. PLoS One. 2015. PMID: 26717567 Free PMC article. - Deacetylase inhibitors repress STAT5-mediated transcription by interfering with bromodomain and extra-terminal (BET) protein function.
Pinz S, Unser S, Buob D, Fischer P, Jobst B, Rascle A. Pinz S, et al. Nucleic Acids Res. 2015 Apr 20;43(7):3524-45. doi: 10.1093/nar/gkv188. Epub 2015 Mar 13. Nucleic Acids Res. 2015. PMID: 25769527 Free PMC article. - Competition-based, quantitative chemical proteomics in breast cancer cells identifies new target profiles for sulforaphane.
Clulow JA, Storck EM, Lanyon-Hogg T, Kalesh KA, Jones LH, Tate EW. Clulow JA, et al. Chem Commun (Camb). 2017 May 4;53(37):5182-5185. doi: 10.1039/c6cc08797c. Chem Commun (Camb). 2017. PMID: 28439590 Free PMC article. - FLT3-ITD confers resistance to the PI3K/Akt pathway inhibitors by protecting the mTOR/4EBP1/Mcl-1 pathway through STAT5 activation in acute myeloid leukemia.
Nogami A, Oshikawa G, Okada K, Fukutake S, Umezawa Y, Nagao T, Kurosu T, Miura O. Nogami A, et al. Oncotarget. 2015 Apr 20;6(11):9189-205. doi: 10.18632/oncotarget.3279. Oncotarget. 2015. PMID: 25826077 Free PMC article.
References
- World Health Organization (WHO) (n.d.) Globocan 2012, Cancer Fact Sheet. Available: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx. Accessed 2014 Mar 12.
- Go VLW, Wong DA, Wang Y, Butrum RR, Norman HA, et al. (2004) Diet and cancer prevention: evidence-based medicine to genomic medicine. J Nutr 134: 3513S–3516S. - PubMed
- Ullrich A, Waxman A, Luiza da Costa e Silva V, Bettcher D, Vestal G, et al. (2004) Cancer prevention in the political arena: the WHO perspective. Ann Oncol Off J Eur Soc Med Oncol ESMO 15 Suppl 4iv249–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by the Deutsche Forschungsgemeinschaft (Grant No. RA 2010/2-1 to AR), the Deutsche Krebshilfe (Grant No. 109750 to AR) and institutional research funds (Foerderlinie C to AR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous