Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex - PubMed (original) (raw)
. 2014 Aug 12;111(4):736-48.
doi: 10.1038/bjc.2014.383. Epub 2014 Jul 15.
L Zhang 2, X Liu 1, L Zhou 1, W Wang 1, Z Han 1, H Sui 1, Y Tang 1, Y Wang 3, N Liu 1, J Ren 4, F Hou 4, Q Li 1
Affiliations
- PMID: 25025966
- PMCID: PMC4134507
- DOI: 10.1038/bjc.2014.383
Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex
Q Ji et al. Br J Cancer. 2014.
Abstract
Background: Metastasis associated with lung adenocarcinoma transcript-1 (MALAT1) is a functional long non-coding RNA (lncRNA), which is highly expressed in several tumours, including colorectal cancer (CRC). Its biological function and mechanism in the prognosis of human CRC is still largely under investigation.
Methods: This study aimed to investigate the new effect mechanism of MALAT1 on the proliferation and migration of CRC cells in vitro and in vivo, and detect the expression of MALAT1, SFPQ (also known as PSF (PTB-associated splicing factor)), and PTBP2 (also known as PTB (polypyrimidine-tract-binding protein)) in CRC tumour tissues, followed by correlated analysis with clinicopathological parameters.
Results: We found that overexpression of MALAT1 could promote cell proliferation and migration in vitro, and promote tumour growth and metastasis in nude mice. The underlying mechanism was associated with tumour suppressor gene SFPQ and proto-oncogene PTBP2. In CRC, MALAT1 could bind to SFPQ, thus releasing PTBP2 from the SFPQ/PTBP2 complex. In turn, the increased SFPQ-detached PTBP2 promoted cell proliferation and migration. SFPQ critically mediated the regulatory effects of MALAT1. Moreover, in CRC tissues, MALAT1 and PTBP2 were overexpressed, both of which were associated closely with the invasion and metastasis of CRC. However, the SFPQ showed unchanged expression either in CRC tissues or adjacent normal tissues.
Conclusions: Our findings implied that MALAT1 might be a potential predictor for tumour metastasis and prognosis. Furthermore, the interaction between MALAT1 and SFPQ could be a novel therapeutic target for CRC.
Figures
Figure 1
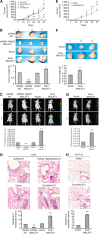
Upregulation and downregulation of MALAT1 expression. (A) Six established human CRC cell lines SW480, HCT116, LoVo, SW620, LS174T, and HCT8 were analysed for the MALAT1 expression by RT–PCR (up) and real-time PCR (down), GAPDH was chosen as a control. (B) RT–PCR was used to amplify three fragments of MALAT1 gene (upper panel), and the full length of MALAT1 gene was spliced by series of PCR (lower panel). (C) LoVo and HCT116 cells were infected with lentiviral particles containing pLV4-over/MALAT1 and pLV4-vector. The MALAT1 transcript was evaluated by RT–PCR (up) and real-time PCR (down). **P<0.01, compared with LoVo-vector or HCT116-vector cells. (D) LoVo cells were infected with lentiviral particles containing pLV4-shRNA/MALAT1 or pLV4-shRNA/NT, and the knockdown efficiency was evaluated by RT–PCR (up) and real-time PCR (down). *P<0.05; **P<0.01, compared with LoVo-shRNA/NT cells.
Figure 2
MALAT1 promoted growth and metastasis of LoVo cells in vivo. (A) LoVo-shRNA/NT, LoVo-shRNA/MALAT1, LoVo-vector, and LoVo-over/MALAT1 cells were respectively injected subcutaneously into nude mice (_n_=8). Length and width of the tumours were measured every 7 days. (B) After 42 days, mice were killed for determination of tumour weights. (C) LoVo-shRNA/NT, LoVo-shRNA/MALAT1, LoVo-vector, and LoVo-over/MALAT1 cells were respectively injected into the lateral tail vein. Seven weeks later, the established lungs metastases images were observed by LB983 NIGHTOWL II system. (D) The organs of lung were excised, the metastases originated from i.v. injections were checked by haematoxylin and eosin (H&E) staining, and the numbers of metastatic lesions were counted. The magnification of the microscopic pictures was × 100, and the selected metastatic lesions in the lower right corner were magnified by × 200. **P<0.01, compared with LoVo-shRNA/NT or LoVo-vector group. (E–H) HCT116-vector and HCT116-over/MALAT1 cells were analysed for the influence of MALAT1 on tumour growth, tumour weights, and metastasis ability in vivo as LoVo cells. *P<0.05; **P<0.01, compared with HCT116-vector group.
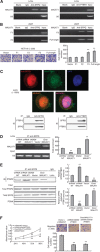
Figure 3
MALAT1 competitively bound to SFPQ and released SFPQ from the SFPQ/PTBP2 complex. (A) RIP analysis between MALAT1 and SFPQ. As a control, mouse monoclonal IgG was used. The blank group was the PCR results with no cDNA, and the input group was the cDNA from cell lysates without RIP procedure. U6, a abundant nuclear RNA, was used as a negative control for enrichment calculation. RIP analysis between MALAT1 and PTBP2 was performed as SFPQ. (B) Four constructed expression vector: pLV4-MALAT1/F1, pLV4-MALAT1/F2, pLV4-MALAT1/F3, and pLV4-over/MALAT1, respectively, together with the pLV4-over/SFPQ vector, were co-transfected into 293T cells, following by RIP analysis, to verify the region of interaction between MALAT1 and SFPQ. Above four expression vectors, together with pLV4-vector, were also transfected into HCT116 cells, and the cell migration was analysed by transwell method. **P<0.01, compared with pLV4-vector group. (C) Immunofluorescence and immunoprecipitation analysis for co-localisation of SFPQ and PTBP2. Protein SFPQ and PTBP2 were respectively detected by Cy3-conjugated secondary antibody (red colour) and FITC-conjugated secondary antibody (green colour), and the nucleus was stained with DAPI (blue colour). The images were taken with a TCS SP2 spectral confocal system. Image MERGE1 was merged with red and green colours, and image MERGE2 was merged with red, green and blue colours. The magnification of the microscopic pictures was × 1000. Immunoprecipitation was applied to verify the results of immunofluorescence. (D) RIP analysis was used to detect the effect of MALAT1 expression on the interaction between MALAT1 and SFPQ in LoVo-shRNA/NT, LoVo-shRNA/MALAT1, LoVo-vector and LoVo-over/MALAT1, and the quantities of the binding were determined by density analysis. **P<0.01, compared with LoVo-shRNA/NT or LoVo-vector group. (E) Extracted proteins from above four cells were immunoprecipitated with anti-SFPQ antibody or mouse IgG control. The precipitates were subjected to western blot with anti-PTBP2 antibody. **P<0.01, compared with LoVo-shRNA/NT or LoVo-vector group. (F) MALAT1 was overexpressed and a simultaneous knockdown of PTBP2 was performed in LoVo cells, and the growth and metastasis were detected and evaluated. **P<0.01, compared with LoVo-shRNA/NT-vector group. The full colour version of this figure is available at British Journal of Cancer online.
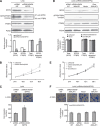
Figure 4
SFPQ played a key role in the regulatory effect of MALAT1 on cell proliferation and migration. (A) LoVo cells were transfected with shRNA/NT or shRNA/SFPQ, and the stable knockdown cells were selected with neomycin. The silencing efficiency of SFPQ was evaluated by western blot. Total quantities of SFPQ/PTBP2 complex were evaluated by immunoprecipitation. Total PTBP2 and control PCNA proteins were detected by western blot. **P<0.01; ##P<0.01; &&P<0.01, compared with LoVo-shRNA/NT cells. (B and C) MTT and transwell assay were performed to evaluate the proliferation and migration ability of LoVo-shRNA/NT and LoVo-shRNA/SFPQ cells. *P<0.05, compared with LoVo-shRNA/NT cells. (D) LoVo-shRNA/SFPQ cells were transiently transfected with siRNA/NT, siRNA/MALAT1, pLV4-vector, and pLV4-over/MALAT1 for 48 h. Total quantities of SFPQ/PTBP2 complex were evaluated by immunoprecipitation. Total SFPQ, PTBP2 and control PCNA proteins were detected by western blot. (E and F) MTT and transwell assay were performed to evaluate the proliferation and migration ability of four kinds of LoVo-shRNA/SFPQ cells transiently transfected with siRNA/NT, siRNA/MALAT1, pLV4-vector, and pLV4-over/MALAT1, respectively.
Figure 5
Role of PTBP2 in the regulation of MALAT1 on the proliferation and migration. (A) LoVo cells were transfected with shRNA/NT or shRNA/PTBP2, and the cells of stable knockdown were selected with neomycin. The silencing efficiency of PTBP2 was evaluated by western blot. Total quantities of SFPQ/PTBP2 complex were evaluated by immunoprecipitation. Total SFPQ and control PCNA proteins were detected by western blot. **P<0.01; ##P<0.01; &&P<0.01, compared with LoVo-shRNA/NT cells. (B and C) MTT and transwell assay were executed to evaluate the proliferation and migration ability of LoVo-shRNA/NT and LoVo-shRNA/PTBP2 cells. **P<0.05, compared with LoVo-shRNA/NT cells. (D) LoVo-shRNA/PTBP2 cells were transiently transfected with siRNA/NT, siRNA/MALAT1, pLV4-vector, and pLV4-over/MALAT1 for 48 h. Total quantities of SFPQ/PTBP2 complex were evaluated by immunoprecipitation. Total SFPQ, PTBP2 and control PCNA proteins were detected by western blot. (E and F) MTT and transwell assay were executed to evaluate the proliferation and migration ability of four kinds of LoVo-shRNA/PTBP2 cells transiently transfected with siRNA/NT, siRNA/MALAT1, pLV4-vector, and pLV4-over/MALAT1, respectively.
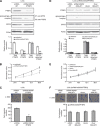
Figure 6
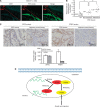
Expression of MALAT1 transcripts, SFPQ and PTBP2 protein in human colorectal cancer, and a hypothetical illustration for the role and interaction of MALAT1, SFPQ and PTBP2. (A) Fluorescence in situ hybridisation was applied to investigate the MALAT1 expression in tissues using both tumour section and normal section. (B) Real-time PCR detected the relative MALAT1 expression level in adjacent normal tissues (_n_=60), CRC tissues with no metastasis (_n_=20, **P<0.01, compared with adjacent normal tissues), CRC tissues with metastasis (_n_=40, **P<0.01, compared with CRC tissues with no metastasis). (C) Immunohistochemistry was performed to detect SFPQ and PTBP2 protein expression in CRC tissues and adjacent normal tissues. Integrated optical densities of each protein expression were determined. **P<0.01, compared with adjacent normal tissues. (D) A hypothetical illustration for the role and interaction of MALAT1, SFPQ and PTBP2.
Similar articles
- MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis.
Ji Q, Cai G, Liu X, Zhang Y, Wang Y, Zhou L, Sui H, Li Q. Ji Q, et al. Cell Death Dis. 2019 May 16;10(6):378. doi: 10.1038/s41419-019-1598-x. Cell Death Dis. 2019. PMID: 31097689 Free PMC article. - MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9.
Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. Yang MH, et al. Biochim Biophys Acta. 2015 Jan;1852(1):166-74. doi: 10.1016/j.bbadis.2014.11.013. Epub 2014 Nov 18. Biochim Biophys Acta. 2015. PMID: 25446987 Free PMC article. - Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO.
Yang P, Chen T, Xu Z, Zhu H, Wang J, He Z. Yang P, et al. Oncotarget. 2016 Jul 5;7(27):42183-42194. doi: 10.18632/oncotarget.9741. Oncotarget. 2016. PMID: 27259250 Free PMC article. - MALAT1: A Promising Therapeutic Target for the Treatment of Metastatic Colorectal Cancer.
Uthman YA, Ibrahim KG, Abubakar B, Bello MB, Malami I, Imam MU, Qusty N, Cruz-Martins N, Batiha GE, Abubakar MB. Uthman YA, et al. Biochem Pharmacol. 2021 Aug;190:114657. doi: 10.1016/j.bcp.2021.114657. Epub 2021 Jun 16. Biochem Pharmacol. 2021. PMID: 34144008 Review. - MALAT1 in colorectal cancer: Its implication as a diagnostic, prognostic, and predictive biomarker.
Cervena K, Vodenkova S, Vymetalkova V. Cervena K, et al. Gene. 2022 Nov 15;843:146791. doi: 10.1016/j.gene.2022.146791. Epub 2022 Aug 9. Gene. 2022. PMID: 35961438 Review.
Cited by
- Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer.
Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Yin DD, et al. Tumour Biol. 2015 Jun;36(6):4851-9. doi: 10.1007/s13277-015-3139-2. Epub 2015 Feb 1. Tumour Biol. 2015. PMID: 25636452 - Long noncoding RNA DGCR5 involves in tumorigenesis of esophageal squamous cell carcinoma via SRSF1-mediated alternative splicing of Mcl-1.
Duan Y, Jia Y, Wang J, Liu T, Cheng Z, Sang M, Lv W, Qin J, Liu L. Duan Y, et al. Cell Death Dis. 2021 Jun 7;12(6):587. doi: 10.1038/s41419-021-03858-7. Cell Death Dis. 2021. PMID: 34099633 Free PMC article. - LncRNAs and Esophageal Squamous Cell Carcinoma - Implications for Pathogenesis and Drug Development.
Shen WJ, Zhang F, Zhao X, Xu J. Shen WJ, et al. J Cancer. 2016 Jun 23;7(10):1258-64. doi: 10.7150/jca.14869. eCollection 2016. J Cancer. 2016. PMID: 27390601 Free PMC article. Review. - Single-Cell Transcriptome Analysis Reveals Paraspeckles Expression in Osteosarcoma Tissues.
Rothzerg E, Feng W, Song D, Li H, Wei Q, Fox A, Wood D, Xu J, Liu Y. Rothzerg E, et al. Cancer Inform. 2022 Dec 5;21:11769351221140101. doi: 10.1177/11769351221140101. eCollection 2022. Cancer Inform. 2022. PMID: 36507075 Free PMC article. - The Intricate Interplay between Epigenetic Events, Alternative Splicing and Noncoding RNA Deregulation in Colorectal Cancer.
Amirkhah R, Naderi-Meshkin H, Shah JS, Dunne PD, Schmitz U. Amirkhah R, et al. Cells. 2019 Aug 19;8(8):929. doi: 10.3390/cells8080929. Cells. 2019. PMID: 31430887 Free PMC article. Review.
References
- Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55 (4:623–631. - PubMed
- Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418 (6900:823. - PubMed
- Chaumeil J, Augui S, Chow JC, Heard E. Combined immunofluorescence, RNA fluorescent in situ hybridization, and DNA fluorescent in situ hybridization to study chromatin changes, transcriptional activity, nuclear organization, and X-chromosome inactivation. Methods Mol Biol. 2008;463:297–308. - PubMed
- Christofori G. New signals from the invasive front. Nature. 2006;441 (7092:444–450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical