Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease - PubMed (original) (raw)
Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease
Edward M Kennedy et al. J Virol. 2014 Oct.
Abstract
High-risk human papillomaviruses (HPVs), including HPV-16 and HPV-18, are the causative agents of cervical carcinomas and are linked to several other tumors of the anogenital and oropharyngeal regions. The majority of HPV-induced tumors contain integrated copies of the normally episomal HPV genome that invariably retain intact forms of the two HPV oncogenes E6 and E7. E6 induces degradation of the cellular tumor suppressor p53, while E7 destabilizes the retinoblastoma (Rb) protein. Previous work has shown that loss of E6 function in cervical cancer cells induces p53 expression as well as downstream effectors that induce apoptosis and cell cycle arrest. Similarly, loss of E7 allows increased Rb expression, leading to cell cycle arrest and senescence. Here, we demonstrate that expression of a bacterial Cas9 RNA-guided endonuclease, together with single guide RNAs (sgRNAs) specific for E6 or E7, is able to induce cleavage of the HPV genome, resulting in the introduction of inactivating deletion and insertion mutations into the E6 or E7 gene. This results in the induction of p53 or Rb, leading to cell cycle arrest and eventual cell death. Both HPV-16- and HPV-18-transformed cells were found to be responsive to targeted HPV genome-specific DNA cleavage. These data provide a proof of principle for the idea that vector-delivered Cas9/sgRNA combinations could represent effective treatment modalities for HPV-induced cancers. Importance: Human papillomaviruses (HPVs) are the causative agents of almost all cervical carcinomas and many other tumors, including many head and neck cancers. In these cancer cells, the HPV DNA genome is integrated into the cellular genome, where it expresses high levels of two viral oncogenes, called E6 and E7, that are required for cancer cell growth and viability. Here, we demonstrate that the recently described bacterial CRISPR/Cas RNA-guided endonuclease can be reprogrammed to target and destroy the E6 or E7 gene in cervical carcinoma cells transformed by HPV, resulting in cell cycle arrest, leading to cancer cell death. We propose that viral vectors designed to deliver E6- and/or E7-specific CRISPR/Cas to tumor cells could represent a novel and highly effective tool to treat and eliminate HPV-induced cancers.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
FIG 1
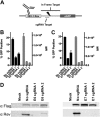
S. pyogenes Cas9 HPV-18-specific sgRNA screening. Two sgRNAs were designed to target the amino terminus of the HPV-18 E6 or E7 protein and screened to identify the most effective candidate. (A) Schematic depicting the fusion protein-based reporter assay, which includes an amino-terminal HIV-1 Rev fragment that acts as an epitope tag, an in-frame HPV-18-derived target sequence, and a carboxy-terminal eGFP ORF (described in detail in Materials and Methods). (B) Graph showing eGFP expression data for 293T cells cotransfected with plasmids expressing the Flag-tagged S. pyogenes Cas9 protein and sgRNAs specific for the HPV-18 E6 gene, or a control construct, and their cognate indicator plasmids. Transfected cells were processed for flow cytometry at 72 h. The number of GFP-positive cells and the mean fluorescence intensity (MFI) of these cells are indicated. Averages and standard deviations of data from three independent experiments are indicated. (C) Similar to panel B but with two HPV-18 E7-specific sgRNAs. (D) Western blot using an HIV-1 Rev-specific antiserum to detect expression of the Rev-GFP indicator fusion protein, thus demonstrating sgRNA efficacy and specificity.
FIG 2
HPV-18 E6- and E7-specific S. pyogenes Cas9 sgRNAs induce mutagenesis at the predicted cleavage site in the HPV genome. (A) E6 and E7 sgRNA and S. pyogenes Cas9 expression constructs were transfected, and the Surveyor assay was performed. The predicted size of the Surveyor cleavage product is indicated by an arrow. DNA markers (left lane) are indicated in base pairs. WT, wild type. (B) Alignment of the DNA sequence of the targeted region of the HPV-18 E6 gene isolated from HeLa cells expressing an E6-specific RGN. The sgRNA target and PAM for the HPV-18 E6 locus are indicated. (C) Similar to panel B but with the HPV-18 E7 gene.
FIG 3
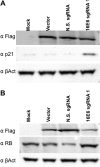
HPV-18 E6- and E7-specific RGNs induce tumor suppressor gene expression in HeLa cells. HeLa cells were transfected with an S. pyogenes Cas9 expression vector and HPV-18-specific sgRNAs, as indicated, and were processed for Western blotting. These data are representative of data from 3 biological replicates. (A) The lysate was probed for p53 and p21 expression, with endogenous β-actin being used as a loading control. The Cas9 protein was detected by an antibody specific for the Flag epitope tag. (B) Similar to panel A except that the lysate was probed for Rb. (C) Sequence of a mutant E6 expression construct designed to be resistant to cleavage by S. pyogenes Cas9 in the presence of E6 sgRNA1. The mutations are in lowercase type, and the PAM is underlined. (D) Expression of the cleavage-resistant E6 gene shown in panel C reveals _trans_-complementation of p53 protein repression in the presence of S. pyogenes Cas9 and E6 sgRNA1. N.S., nonspecific.
FIG 4
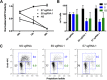
RGN-directed mutagenesis of either the E6 or E7 locus induces cell cycle arrest in G1. (A) S. pyogenes Cas9-mediated disruption of either the HPV-18 E6 or E7 gene results in the expected inhibition of HeLa cell growth. The number of GFP-positive cells transfected with vectors encoding S. pyogenes Cas9, a control or HPV-specific sgRNA, and the gfp gene is shown. (B) Cell cycle analysis of HeLa cells expressing E6- or E7-specific RGNs using BrdU incorporation and PI staining followed by fluorescence-activated cell sorter analysis. Results from four separate biological replicates of the transfected eGFP-positive HeLa cell population are shown. (C) Representative flow cytometry plots are shown for the sgRNAs indicated. The percentage of GFP-positive HeLa cells in each phase of the cell cycle was quantitated and is indicated. NS, nonspecific; APC, allophycocyanin.
FIG 5
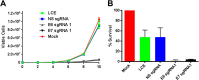
Lentiviral vectors expressing S. pyogenes Cas9 and sgRNAs specific for the HPV-18 E6 and E7 genes induce the death of cervical carcinoma cells. HeLa cells were transduced with a lentiviral vector expressing eGFP, to control for lentiviral toxicity (LCE), or a lentiviral vector expressing S. pyogenes Cas9 and a nonspecific sgRNA or E6- or E7-specific sgRNAs. (A) HeLa cells were transduced with the lentiviral vector at an MOI of ∼2.2, resulting in transduction of ∼90% of the cells in culture. Cell growth was then monitored over a 10-day period. Data from a representative experiment, derived from three similar independent experiments, are presented. (B) HeLa cells were transduced with the lentiviral vector at an MOI of ∼37, which is predicted to transduce >99% of the cells in culture. The cells were then assayed for viability by MTT staining on day 10. Averages of data from two independent experiments normalized to values for a mock-transduced culture are shown. N.S., nonspecific.
FIG 6
HPV-16 E6 and E7 RGNs rescue p21 and Rb expression in SiHa cells. SiHa cells were transfected with an S. pyogenes Cas9 expression vector and the HPV-16-specific sgRNA constructs indicated and processed for Western blotting. These data are representative of data from 3 biological replicates. (A) Lysate probed for the Flag-tagged Cas9 protein, the p53 effector protein p21, and endogenous β-actin (B). Similar to panel A but with the lysate being probed for Rb expression. The N.S. (nonspecific) sgRNA used as a control is E6 sgRNA1, which is specific for the HPV-18 E6 gene but is not predicted to recognize the HPV-16 E6 gene.
Similar articles
- In vitro and in vivo growth inhibition of human cervical cancer cells via human papillomavirus E6/E7 mRNAs' cleavage by CRISPR/Cas13a system.
Chen Y, Jiang H, Wang T, He D, Tian R, Cui Z, Tian X, Gao Q, Ma X, Yang J, Wu J, Tan S, Xu H, Tang X, Wang Y, Yu Z, Han H, Das BC, Severinov K, Hitzeroth II, Debata PR, Xu W, Fan W, Jin Z, Cao C, Yu M, Xie W, Huang Z, Hu Z, You Z. Chen Y, et al. Antiviral Res. 2020 Jun;178:104794. doi: 10.1016/j.antiviral.2020.104794. Epub 2020 Apr 14. Antiviral Res. 2020. PMID: 32298665 - Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways.
Goodwin EC, DiMaio D. Goodwin EC, et al. Proc Natl Acad Sci U S A. 2000 Nov 7;97(23):12513-8. doi: 10.1073/pnas.97.23.12513. Proc Natl Acad Sci U S A. 2000. PMID: 11070078 Free PMC article. - High-Risk Alphapapillomavirus Oncogenes Impair the Homologous Recombination Pathway.
Wallace NA, Khanal S, Robinson KL, Wendel SO, Messer JJ, Galloway DA. Wallace NA, et al. J Virol. 2017 Sep 27;91(20):e01084-17. doi: 10.1128/JVI.01084-17. Print 2017 Oct 15. J Virol. 2017. PMID: 28768872 Free PMC article. - Genomic Signatures in HPV-Associated Tumors.
Hussain SS, Lundine D, Leeman JE, Higginson DS. Hussain SS, et al. Viruses. 2021 Oct 5;13(10):1998. doi: 10.3390/v13101998. Viruses. 2021. PMID: 34696429 Free PMC article. Review. - Molecular mechanism of carcinogenesis by human papillomavirus-16.
Ishiji T. Ishiji T. J Dermatol. 2000 Feb;27(2):73-86. doi: 10.1111/j.1346-8138.2000.tb02126.x. J Dermatol. 2000. PMID: 10721654 Review.
Cited by
- CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy.
Xu Y, Li Z. Xu Y, et al. Comput Struct Biotechnol J. 2020 Sep 8;18:2401-2415. doi: 10.1016/j.csbj.2020.08.031. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 33005303 Free PMC article. Review. - Non-human primate papillomavirus E6-mediated p53 degradation reveals ancient evolutionary adaptation of carcinogenic phenotype to host niche.
Long T, Burk RD, Chan PKS, Chen Z. Long T, et al. PLoS Pathog. 2022 Mar 25;18(3):e1010444. doi: 10.1371/journal.ppat.1010444. eCollection 2022 Mar. PLoS Pathog. 2022. PMID: 35333912 Free PMC article. - HPV Oncogene Manipulation Using Nonvirally Delivered CRISPR/Cas9 or Natronobacterium gregoryi Argonaute.
Lao YH, Li M, Gao MA, Shao D, Chi CW, Huang D, Chakraborty S, Ho TC, Jiang W, Wang HX, Wang S, Leong KW. Lao YH, et al. Adv Sci (Weinh). 2018 May 18;5(7):1700540. doi: 10.1002/advs.201700540. eCollection 2018 Jul. Adv Sci (Weinh). 2018. PMID: 30027026 Free PMC article. - Sharpening the Molecular Scissors: Advances in Gene-Editing Technology.
Broeders M, Herrero-Hernandez P, Ernst MPT, van der Ploeg AT, Pijnappel WWMP. Broeders M, et al. iScience. 2020 Jan 24;23(1):100789. doi: 10.1016/j.isci.2019.100789. Epub 2019 Dec 19. iScience. 2020. PMID: 31901636 Free PMC article. Review. - CRISPR/Cas9-Based Antiviral Strategy: Current Status and the Potential Challenge.
Lee C. Lee C. Molecules. 2019 Apr 5;24(7):1349. doi: 10.3390/molecules24071349. Molecules. 2019. PMID: 30959782 Free PMC article. Review.
References
- Howley PM. 1991. Role of the human papillomaviruses in human cancer. Cancer Res. 51:5019s–5022s - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous