Molecular and cellular insights into T cell exhaustion - PubMed (original) (raw)
Review
Molecular and cellular insights into T cell exhaustion
E John Wherry et al. Nat Rev Immunol. 2015 Aug.
Abstract
In chronic infections and cancer, T cells are exposed to persistent antigen and/or inflammatory signals. This scenario is often associated with the deterioration of T cell function: a state called 'exhaustion'. Exhausted T cells lose robust effector functions, express multiple inhibitory receptors and are defined by an altered transcriptional programme. T cell exhaustion is often associated with inefficient control of persisting infections and tumours, but revitalization of exhausted T cells can reinvigorate immunity. Here, we review recent advances that provide a clearer molecular understanding of T cell exhaustion and reveal new therapeutic targets for persisting infections and cancer.
Figures
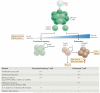
Figure 1. Progressive development of T cell exhaustion
Upon infection, naive T cells are activated by antigen, co-stimulation and inflammation, and they exponentially proliferate to form effector populations,. Whereas the majority of effector CD8+ T cells that express killer cell lectin-like receptor subfamily G member 1 (KLRG1) die during the contraction phase, a population of effector CD8+ T cells that retains CD127 expression can give rise to memory or exhausted CD8+ T cells. In the setting of acute infection, where antigen and/or inflammation is cleared, effector CD8+ T cells further differentiate into functional memory CD8+ T cells that can produce multiple cytokines (such as interferon-γ (IFNγ), tumour necrosis factor (TNF) and interleukin-2 (IL-2)) and mount robust recall responses upon secondary infection,. These memory T cells are also maintained efficiently long term without antigen via IL-7- and IL-15-driven homeostatic self-renewal. By contrast, during chronic infection, antigen and inflammation persist after the effector phase. As infection progresses and T cell stimulation continues, T cells lose effector functions in a hierarchical manner and become exhausted. Typically, functions such as IL-2 production and cytokine polyfunctionality, as well as high proliferative capacity, are lost early; this is followed by defects in the production of IFNγ, TNF and chemokines, as well as in degranulation. T cell exhaustion is also accompanied by a progressive increase in the amount and diversity of inhibitory receptors that are expressed, including programmed cell death protein 1 (PD1), lymphocyte activation gene 3 protein (LAG3), 2B4, CD160 and T cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT). Ultimately, if the severity or duration of the infection is high or prolonged, virus-specific T cells can be lost (‘deletion’). Variables — such as the level and number of inhibitory receptors expressed, strength of antigen stimulation, availability of CD4+ T cell help and the duration of infection — can all influence the severity of exhaustion. Exhausted T cell populations are heterogeneous, and subsets of T-bethi PD1mid and EOMEShi PD1hi CD8+ exhausted T cells exist. Only the T-bethi PD1mid subset is responsive to reinvigoration by blockade of the PD1 pathway. CXCR3, CXC-chemokine receptor 3; PDL1, PD1 ligand 1.
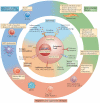
Figure 2. Overview of mechanisms of T cell exhaustion
Pathways implicated in regulating T cell exhaustion can be classified into three general categories (centre and inner circle): cell-to-cell signals including prolonged T cell receptor (TCR) engagement (signal 1) and co-stimulatory and/or co-inhibitory signals (signal 2); soluble factors such as excessive levels of inflammatory cytokines (for example, type I interferons (IFNs)) and suppressive cytokines including interleukin-10 (IL-10) and transforming growth factor-β (TGFβ); and tissue and microenvironmental influences driven by changes in the expression levels of chemokine receptors, adhesion molecules and nutrient receptors. This last class of influences may include altered tissue distribution and/or migratory patterns and lead to changes in pathways sensing oxygen tension (the von Hippel–Lindau tumour suppressor (VHL) and/or hypoxia-inducible factor (HIF) pathways), pH and nutrient levels. Tissue destruction and altered lymphoid organization may have a major role. Other immune cell types and stromal cells could be the source of many of these changes (outer circle). Cell types such as antigen-presenting cells (APCs), CD4+ T cells, natural killer (NK) cells, B cells and regulatory cells (for example, myeloid-derived suppressor cells (MDSCs) and regulatory T (TReg) cells) have been implicated in CD8+ T cell exhaustion. Overall, during chronic infections, cell-intrinsic and cell-extrinsic signals are probably integrated and thereby negatively influence T cell differentiation and promote exhaustion. The precise balance of these signals may determine the severity and/or qualitative aspects of T cell exhaustion in different disease settings. CTLA4, cytotoxic T lymphocyte antigen 4; DC, dendritic cell; FOXP3, forkhead box P3; LAG3, lymphocyte activation gene 3 protein; PD1, programmed cell death protein 1; TH cell, T helper cell.
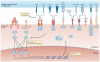
Figure 3. Molecular pathways of inhibitory receptors associated with T cell exhaustion
Ligand and receptor pairs for inhibitory pathways are depicted, showing the intracellular domains of receptors that contribute to T cell exhaustion. Many inhibitory receptors have immunoreceptor tyrosine-based inhibitory motifs (ITIMs) and/or immunoreceptor tyrosine-based switch motifs (ITSMs) in their intracellular domains; however, some receptors have specific motifs, such as YVKM for cytotoxic T lymphocyte antigen 4 (CTLA4) and KIEELE for lymphocyte activation gene 3 protein (LAG3). The molecular mechanisms of inhibitory receptor signalling are also illustrated and can be classified as: ectodomain competition (inhibitory receptors sequester target receptors or ligands); modulation of intracellular mediators (local and transient intracellular attenuation of positive signals from activating receptors such as T cell receptors and co-stimulatory receptors); and induction of inhibitory genes. Multiple inhibitory receptors are responsible for these three mechanisms. AP-1, activator protein 1; BAT3, HLA-B-associated transcript 3 (also known as BAG6); BTLA, B and T lymphocyte attenuator; CEACAM1, carcinoembryonic antigen-related cell adhesion molecule 1; GRB2, growth factor receptor-bound protein 2; HVEM, herpes virus entry mediator (also known as TNFRSF14); NFAT, nuclear factor of activated T cells; NF-κB, nuclear factor-κB; PD1, programmed cell death protein 1; PDL1, PD1 ligand 1; PI3K, phosphoinositide 3-kinase; PLCγ, phospholipase Cγ; TIGIT, T cell immunoreceptor with immunoglobulin and ITIM domains; TIM3, T cell immunoglobulin and mucin domain-containing protein 3.
Figure 4. Transcriptional and epigenetic mechanisms of T cell exhaustion
Altered usage of key transcription factors is associated with the altered transcriptional and developmental programme of T cell exhaustion. Several potential mechanisms exist. a | Different use of transcription factor binding partners is one potential mechanism for distinct context-dependent transcription factor activity. In the example shown, transcription factor B is closely associated with memory-related genes in the context of acute infection where transcription factor A is abundant (top panel). However, the same transcription factor B is linked with exhaustion-related genes in chronic infection (lower panel). Post-translational modifications of transcription factors and/or their subcellular localization may also be important in this setting. b | The dosage or concentration of a transcription factor could also provide a mechanism for context-dependent transcriptional function. Here, a transcription factor binds only at specific high-affinity binding sites if the amount of the factor is low (top panel). By contrast, at high transcription factor concentrations binding can occur more broadly (that is, occurring also at lower affinity sites), which leads to different transcriptional activity (lower panel). c | DNA methylation, histone modifications and the ‘chromatin landscape’ resulting from overall epigenetic regulation could provide a mechanism for context-specific transcription factor function in exhausted T cells (transcription factor X in this example). The enhancer landscape of a cell may determine how different transcription factors function.
Similar articles
- Positive and negative regulation of cellular immune responses in physiologic conditions and diseases.
Viganò S, Perreau M, Pantaleo G, Harari A. Viganò S, et al. Clin Dev Immunol. 2012;2012:485781. doi: 10.1155/2012/485781. Epub 2012 Mar 26. Clin Dev Immunol. 2012. PMID: 22548114 Free PMC article. Review. - Exhaustion of T lymphocytes in the tumor microenvironment: Significance and effective mechanisms.
Davoodzadeh Gholami M, Kardar GA, Saeedi Y, Heydari S, Garssen J, Falak R. Davoodzadeh Gholami M, et al. Cell Immunol. 2017 Dec;322:1-14. doi: 10.1016/j.cellimm.2017.10.002. Epub 2017 Oct 10. Cell Immunol. 2017. PMID: 29079339 Review. - T cell exhaustion.
Wherry EJ. Wherry EJ. Nat Immunol. 2011 Jun;12(6):492-9. doi: 10.1038/ni.2035. Nat Immunol. 2011. PMID: 21739672 Review. - T cell exhaustion during persistent viral infections.
Kahan SM, Wherry EJ, Zajac AJ. Kahan SM, et al. Virology. 2015 May;479-480:180-93. doi: 10.1016/j.virol.2014.12.033. Epub 2015 Jan 22. Virology. 2015. PMID: 25620767 Free PMC article. Review. - Regulatory T Cells and Cancer: A Two-Sided Story.
Wang K, Vella AT. Wang K, et al. Immunol Invest. 2016 Nov;45(8):797-812. doi: 10.1080/08820139.2016.1197242. Epub 2016 Sep 7. Immunol Invest. 2016. PMID: 27603750 Review.
Cited by
- Enhancing cellular immunotherapies in cancer by engineering selective therapeutic resistance.
Wellhausen N, Baek J, Gill SI, June CH. Wellhausen N, et al. Nat Rev Cancer. 2024 Sep;24(9):614-628. doi: 10.1038/s41568-024-00723-5. Epub 2024 Jul 24. Nat Rev Cancer. 2024. PMID: 39048767 Review. - Pre-exposure to mRNA-LNP inhibits adaptive immune responses and alters innate immune fitness in an inheritable fashion.
Qin Z, Bouteau A, Herbst C, Igyártó BZ. Qin Z, et al. bioRxiv [Preprint]. 2022 Aug 20:2022.03.16.484616. doi: 10.1101/2022.03.16.484616. bioRxiv. 2022. PMID: 36032972 Free PMC article. Updated. Preprint. - Identification of immunological subtypes of hepatocellular carcinoma with expression profiling of immune-modulating genes.
Cao D, Chen MK, Zhang QF, Zhou YF, Zhang MY, Mai SJ, Zhang YJ, Chen MS, Li XX, Wang HY. Cao D, et al. Aging (Albany NY). 2020 Jun 16;12(12):12187-12205. doi: 10.18632/aging.103395. Epub 2020 Jun 16. Aging (Albany NY). 2020. PMID: 32544882 Free PMC article. - Understanding the Immune-Stroma Microenvironment in B Cell Malignancies for Effective Immunotherapy.
Apollonio B, Ioannou N, Papazoglou D, Ramsay AG. Apollonio B, et al. Front Oncol. 2021 Mar 25;11:626818. doi: 10.3389/fonc.2021.626818. eCollection 2021. Front Oncol. 2021. PMID: 33842331 Free PMC article. Review. - Microanatomy of the Human Atherosclerotic Plaque by Single-Cell Transcriptomics.
Depuydt MAC, Prange KHM, Slenders L, Örd T, Elbersen D, Boltjes A, de Jager SCA, Asselbergs FW, de Borst GJ, Aavik E, Lönnberg T, Lutgens E, Glass CK, den Ruijter HM, Kaikkonen MU, Bot I, Slütter B, van der Laan SW, Yla-Herttuala S, Mokry M, Kuiper J, de Winther MPJ, Pasterkamp G. Depuydt MAC, et al. Circ Res. 2020 Nov 6;127(11):1437-1455. doi: 10.1161/CIRCRESAHA.120.316770. Epub 2020 Sep 28. Circ Res. 2020. PMID: 32981416 Free PMC article.
References
- Masopust D, Schenkel JM. The integration of T cell migration, differentiation and function. Nat. Rev. Immunol. 2013;13:309–320. - PubMed
- Wherry EJ. T cell exhaustion. Nat. Immunol. 2011;131:492–499. - PubMed
- Doering TA, et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–1144. This study shows that memory and exhausted CD8+ T cells have partially non-overlapping modules and centrally connected genes that are thought to be the hubs or foci of biological processes. This reference also indicates that transcription factors have distinct connections in exhausted CD8+ T cells compared with memory CD8+ T cells. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI105343/AI/NIAID NIH HHS/United States
- AI112521/AI/NIAID NIH HHS/United States
- AI082630/AI/NIAID NIH HHS/United States
- U19 AI082630/AI/NIAID NIH HHS/United States
- P01 AI112521/AI/NIAID NIH HHS/United States
- AI095608/AI/NIAID NIH HHS/United States
- HHSN266200500030C/PHS HHS/United States
- R01 AI105343/AI/NIAID NIH HHS/United States
- U01 AI095608/AI/NIAID NIH HHS/United States
- HHSN266200500030C/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical