Automatic Generation of Validated Specific Epitope Sets - PubMed (original) (raw)
Automatic Generation of Validated Specific Epitope Sets
Sebastian Carrasco Pro et al. J Immunol Res. 2015.
Abstract
Accurate measurement of B and T cell responses is a valuable tool to study autoimmunity, allergies, immunity to pathogens, and host-pathogen interactions and assist in the design and evaluation of T cell vaccines and immunotherapies. In this context, it is desirable to elucidate a method to select validated reference sets of epitopes to allow detection of T and B cells. However, the ever-growing information contained in the Immune Epitope Database (IEDB) and the differences in quality and subjects studied between epitope assays make this task complicated. In this study, we develop a novel method to automatically select reference epitope sets according to a categorization system employed by the IEDB. From the sets generated, three epitope sets (EBV, mycobacteria and dengue) were experimentally validated by detection of T cell reactivity ex vivo from human donors. Furthermore, a web application that will potentially be implemented in the IEDB was created to allow users the capacity to generate customized epitope sets.
Figures
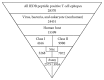
Figure 1
Diagram of the filtering steps towards the generation of the validated sets of epitopes, including the number of epitopes found in each step.
Figure 2
(a) Web application main page interface. (b) Web application “advanced options” page interface.
Figure 3
Predicted epitope pools induce a detectable ex vivo cytokine response. PBMC from donors with LTBI (MTB donor) or previously exposed to DENV were stimulated with their respective epitope pools and an EBV epitope pool. After ex vivo stimulation, the IFN_γ_ response was measured by ICS using flow cytometry. Gating strategy (a) and representative plots of the responses for both CD4+ and CD8+ T cells are shown for a MTB donor (b) and DENV donor (c). (d) The ICS IFN_γ_ responses induced by each epitope pool for all donors tested are summarized (n = 10). Statistical difference was determined using a one-tailed Mann-Whitney test. (∗ P ≤ 0.05; ∗∗ P ≤ 0.01).
Similar articles
- An in vitro-transcribed-mRNA polyepitope construct encoding 32 distinct HLA class I-restricted epitopes from CMV, EBV, and Influenza for use as a functional control in human immune monitoring studies.
Nielsen JS, Wick DA, Tran E, Nelson BH, Webb JR. Nielsen JS, et al. J Immunol Methods. 2010 Aug 31;360(1-2):149-56. doi: 10.1016/j.jim.2010.07.003. Epub 2010 Jul 15. J Immunol Methods. 2010. PMID: 20637775 - Fundamentals and Methods for T- and B-Cell Epitope Prediction.
Sanchez-Trincado JL, Gomez-Perosanz M, Reche PA. Sanchez-Trincado JL, et al. J Immunol Res. 2017;2017:2680160. doi: 10.1155/2017/2680160. Epub 2017 Dec 28. J Immunol Res. 2017. PMID: 29445754 Free PMC article. Review. - Epitope specific T-cell responses against influenza A in a healthy population.
Savic M, Dembinski JL, Kim Y, Tunheim G, Cox RJ, Oftung F, Peters B, Mjaaland S. Savic M, et al. Immunology. 2016 Feb;147(2):165-77. doi: 10.1111/imm.12548. Epub 2015 Dec 8. Immunology. 2016. PMID: 26489873 Free PMC article. - Strategies to query and display allergy-derived epitope data from the immune epitope database.
Vaughan K, Peters B, Larche M, Pomes A, Broide D, Sette A. Vaughan K, et al. Int Arch Allergy Immunol. 2013;160(4):334-45. doi: 10.1159/000343880. Epub 2012 Nov 21. Int Arch Allergy Immunol. 2013. PMID: 23172234 Free PMC article. Review.
Cited by
- HLA-DRB1 Alleles Are Associated With Different Magnitudes of Dengue Virus-Specific CD4+ T-Cell Responses.
Weiskopf D, Angelo MA, Grifoni A, O'Rourke PH, Sidney J, Paul S, De Silva AD, Phillips E, Mallal S, Premawansa S, Premawansa G, Wijewickrama A, Peters B, Sette A. Weiskopf D, et al. J Infect Dis. 2016 Oct 1;214(7):1117-24. doi: 10.1093/infdis/jiw309. Epub 2016 Jul 20. J Infect Dis. 2016. PMID: 27443615 Free PMC article. - SARS-CoV-2 Omicron variant escapes neutralizing antibodies and T cell responses more efficiently than other variants in mild COVID-19 convalescents.
Garcia-Valtanen P, Hope CM, Masavuli MG, Yeow AEL, Balachandran H, Mekonnen ZA, Al-Delfi Z, Abayasingam A, Agapiou D, Stella AO, Aggarwal A, Bouras G, Gummow J, Ferguson C, O'Connor S, McCartney EM, Lynn DJ, Maddern G, Gowans EJ, Reddi BAJ, Shaw D, Kok-Lim C, Beard MR, Weiskopf D, Sette A, Turville SG, Bull RA, Barry SC, Grubor-Bauk B. Garcia-Valtanen P, et al. Cell Rep Med. 2022 Jun 21;3(6):100651. doi: 10.1016/j.xcrm.2022.100651. Epub 2022 May 17. Cell Rep Med. 2022. PMID: 35654046 Free PMC article. - Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals.
Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A. Grifoni A, et al. Cell. 2020 Jun 25;181(7):1489-1501.e15. doi: 10.1016/j.cell.2020.05.015. Epub 2020 May 20. Cell. 2020. PMID: 32473127 Free PMC article. - Urinary Peptides As a Novel Source of T Cell Allergen Epitopes.
da Silva Antunes R, Pham J, McMurtrey C, Hildebrand WH, Phillips E, Mallal S, Sidney J, Busse P, Peters B, Schulten V, Sette A. da Silva Antunes R, et al. Front Immunol. 2018 Apr 26;9:886. doi: 10.3389/fimmu.2018.00886. eCollection 2018. Front Immunol. 2018. PMID: 29755469 Free PMC article. - Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans.
Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, Burger ZC, Rawlings SA, Smith DM, Phillips E, Mallal S, Lammers M, Rubiro P, Quiambao L, Sutherland A, Yu ED, da Silva Antunes R, Greenbaum J, Frazier A, Markmann AJ, Premkumar L, de Silva A, Peters B, Crotty S, Sette A, Weiskopf D. Mateus J, et al. Science. 2020 Oct 2;370(6512):89-94. doi: 10.1126/science.abd3871. Epub 2020 Aug 4. Science. 2020. PMID: 32753554 Free PMC article.
References
- Andrieu J.-M., Chen S., Lai C., Guo W., Lu W. Mucosal SIV vaccines comprising inactivated virus particles and bacterial adjuvants induce CD8+ T-regulatory cells that suppress SIV-positive CD4+ T-cell activation and prevent SIV infection in the macaque model. Frontiers in Immunology. 2014;5, article 297 doi: 10.3389/fimmu.2014.00297. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSN272200900044C/AI/NIAID NIH HHS/United States
- U19 AI100275/AI/NIAID NIH HHS/United States
- HHSN272201200010C/AI/NIAID NIH HHS/United States
- HHSN272201200010C/PHS HHS/United States
- HHSN272200900044C/PHS HHS/United States
- HHSN27220140045C/PHS HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources