Personalized Preclinical Trials in BRAF Inhibitor-Resistant Patient-Derived Xenograft Models Identify Second-Line Combination Therapies - PubMed (original) (raw)
. 2016 Apr 1;22(7):1592-602.
doi: 10.1158/1078-0432.CCR-15-1762. Epub 2015 Dec 16.
Min Xiao 2, Katrin Sproesser 2, Patricia A Brafford 2, Batool Shannan 2, Marilda Beqiri 2, Qin Liu 2, Wei Xu 3, Bradley Garman 3, Katherine L Nathanson 3, Xiaowei Xu 3, Giorgos C Karakousis 3, Gordon B Mills 4, Yiling Lu 4, Tamer A Ahmed 5, Poulikos I Poulikakos 5, Giordano Caponigro 6, Markus Boehm 6, Malte Peters 6, Lynn M Schuchter 3, Ashani T Weeraratna 2, Meenhard Herlyn 2
Affiliations
- PMID: 26673799
- PMCID: PMC4818716
- DOI: 10.1158/1078-0432.CCR-15-1762
Personalized Preclinical Trials in BRAF Inhibitor-Resistant Patient-Derived Xenograft Models Identify Second-Line Combination Therapies
Clemens Krepler et al. Clin Cancer Res. 2016.
Abstract
Purpose: To test second-line personalized medicine combination therapies, based on genomic and proteomic data, in patient-derived xenograft (PDX) models.
Experimental design: We established 12 PDXs from BRAF inhibitor-progressed melanoma patients. Following expansion, PDXs were analyzed using targeted sequencing and reverse-phase protein arrays. By using multi-arm preclinical trial designs, we identified efficacious precision medicine approaches.
Results: We identified alterations previously described as drivers of resistance: NRAS mutations in 3 PDXs, MAP2K1 (MEK1) mutations in 2, BRAF amplification in 4, and aberrant PTEN in 7. At the protein level, re-activation of phospho-MAPK predominated, with parallel activation of PI3K in a subset. Second-line efficacy of the pan-PI3K inhibitor BKM120 with either BRAF (encorafenib)/MEK (binimetinib) inhibitor combination or the ERK inhibitor VX-11e was confirmed in vivo Amplification of MET was observed in 3 PDX models, a higher frequency than expected and a possible novel mechanism of resistance. Importantly, MET amplification alone did not predict sensitivity to the MET inhibitor capmatinib. In contrast, capmatinib as single agent resulted in significant but transient tumor regression in a PDX with resistance to BRAF/MEK combination therapy and high pMET. The triple combination capmatinib/encorafenib/binimetinib resulted in complete and sustained tumor regression in all animals.
Conclusions: Genomic and proteomic data integration identifies dual-core pathway inhibition as well as MET as combinatorial targets. These studies provide evidence for biomarker development to appropriately select personalized therapies of patients and avoid treatment failures. See related commentary by Hartsough and Aplin, p. 1550.
©2015 American Association for Cancer Research.
Conflict of interest statement
Potential conflicts of interest: Part of the studies was supported by a research grant from Novartis.
Figures
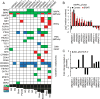
Figure 1. establishment of 12 PDX from BRAF inhibitor progressed patients
A Schematic of BRAF inhibitor resistant PDX generation, expansion, characterization, and in vivo testing. The arrow shown in blue outline denotes possible future clinical translation. B. Patient's progression free survival (PFS) is measured from first day on drug (vemurafenib/solid bar or dabrafenib/hatched bar) to progression by RECIST. Best response is denoted by the color of each bar: SD- light blue, PR- blue, surgical CR- green. Each bar represents one patient, 1/2 denote two samples collected from the same patient. C. PDX tumor grafts are followed from implantation until palpable, blue bars are untreated mice, red bars are mice continuously dosed with BRAF inhibitor (PLX4720) diet. Mean of mouse cohorts (n>5) is shown, error bars are SEM. D. the growth rate of the same tumor grafts from palpable until sacrifice is calculated by max tumor volume in mm3/time in weeks, thus higher values indicate a faster growth rate; blue bars are untreated mice, red bars are mice continuously dosed with BRAF inhibitor (PLX4720) diet. Mean of mouse cohorts (n>5) is shown, error bars are SEM. E. Histology of patient tumor tissue used to generate the PDX and subsequent PDX passages harvested;WM3965-2 is shown as a representative model, all others are shown in supplementary figure S2; MP1: first mouse passage; MP4: 4th serial transplantation in mice; on BRAFi: mice were continuously dosed with BRAF inhibitor diet; H&E staining except PDX S100, a melanoma marker.
Figure 2. targeted sequencing to identify targetable alterations
A DNA alterations identified in 12 PDX samples. Only known somatic short-variants, deletions and amplifications in found in at least one PDX are shown; the full data set of alterations in343 exons and introns can be found in supplementary figure S3 as an excel file. PDX sorted by number of concomitant alterations are in columns, genes sorted by biological pathway are in rows. Mutations are in green, amplifications in red, deletions in blue, and black squares indicate the number of concomitant alterations. B. Levels of ERK/MAPK protein phosphorylation, measured by RPPA, as a surrogate for MAPK pathway activation; BRAFi mice were under continuous BRAF inhibitor therapy at time of harvest, mean of 3 biological replicates is shown, error bars are SD. C. Fold change in AKT phosphorylation between untreated and BRAF inhibitor treated tumors as a surrogate for PI3K pathway re-activation under therapy, mean of 3 biological replicates is shown.
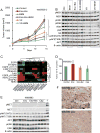
Figure 3. dual pathway inhibition controls tumor growth
A tumor growth curves of WM3936-2 PDX. Animals were treated with vehicle control, encorafenib 20mg/kg QD+binimetinib 3mg/kg QD (Enc+Bin), BKM120 30mg/kg QD, the triple combination encorafenib+binimetinib+BKM120, VX-11e 50mg/kg BID (VX), or VX-11e+BKM120. Dosing was started with established tumors, all compounds were administered orally, n=10/group, error bars are SEM, * indicates p<0.0001. B. immunoblot of tumors harvested at end of study, 4 hours post last dose. The membrane was probed with indicated antibodies. B-Actin was included to ensure equal loading. C. levels of RTK proteins assessed by RPPA, red higher than median, green lower than median, unsupervised hierarchical clustering, data is mean of 3 biological replicates. D. relative tumor growth (final volume/days to max volume) of WM3983 PDX relative to vehicle control. In two separate experiments, animals were treated with 1) vehicle control, encorafenib 20mg/kg QD, encorafenib+binimetinib 3mg/kg QD and 2) vehicle control, capmatinib 25mg/kg QD (Cap). In both experiments, dosing was started with established tumors. All compounds were administered orally for 14 days, n=10/group, error bars are SEM. E. immunoblot of tumor grafts harvested after 3 days of dosing 4 hours post last dose. The membrane was probed with indicated antibodies. B-Actin was included to ensure equal loading.+ denotes the WM3965 tumor graft tissue included as a positive control with elevated levels of MET and pMET. F. IHC staining for MET of patient's melanoma lesion. The pre BRAF inhibitor biopsy shows a strong membrane stain for MET, but only in a subpopulation. The post progression biopsy is negative for MET, the positive cells are macrophages (this lesion was used to establish the PDX).
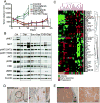
Figure 4. integrating genomic and protein signaling results in an effective triple therapy pre-clinical in vivo trial
A tumor growth curves of WM3965 PDX (MET amplified, high pMET);dosing was started with well-established tumors expanded on BRAF inhibitor diet followed by a washout period before start of dosing. Animals received either vehicle control, encorafenib 20mg/kg QD, binimetinib 3mg/kg QD, capmatinib 25mg/kg QD as single agents or combinations as indicated. All compounds were administered orally, n=10/group, error bars are SEM, * indicates p<0.05. B. immunoblot of tumors harvested after 3 days of dosing and 4 hours post last dose. The membrane was probed with indicated antibodies and B-Actin was included to ensure equal loading. C. RPPA analysis of tumors harvested either at the end of the efficacy experiments (A) or 3 days of dosing (B). Mice without palpable tumors were not included (all triple combo animals). Color coding on the lower x-axis denotes the following groups: blue are control animals (vehicle), orange are the 6animals treated with capmatinib orencorafenib/capmatinib for 3 days (early responders), and pink were treated with capmatinib or encorafenib/capmatinib in the efficacy cohorts and progressed after initial response (progression). Proteins with similar expression along all samples were excluded. The proteins that vary across the samples over a cutoff of 0.4 standard deviations are shown on the y axis. Green is down, red is up regulated. Unsupervised hierarchical clustering of the log2 median centered data is shown on the top x axis. D. MET IHC staining (brown) of FFPE patient tissue. The pre BRAF inhibitor sample is the safety margin around the primary lesion and shows residual melanomanests in the dermal layer (black circle). E. a lymph node metastasis of the same patient after progression (this biopsy was used to establish the PDX).
Comment in
- Of Mice and Melanoma: PDX System for Modeling Personalized Medicine.
Hartsough EJ, Aplin AE. Hartsough EJ, et al. Clin Cancer Res. 2016 Apr 1;22(7):1550-2. doi: 10.1158/1078-0432.CCR-15-3054. Epub 2016 Feb 3. Clin Cancer Res. 2016. PMID: 26842234 Free PMC article. - Using avatars to win the fight over BRAF inhibitor resistance.
Vilgelm AE, Richmond A. Vilgelm AE, et al. Pigment Cell Melanoma Res. 2016 Jul;29(4):398-9. doi: 10.1111/pcmr.12473. Epub 2016 May 17. Pigment Cell Melanoma Res. 2016. PMID: 27185579 Free PMC article. No abstract available.
Similar articles
- Dual MAPK Inhibition Is an Effective Therapeutic Strategy for a Subset of Class II BRAF Mutant Melanomas.
Dankner M, Lajoie M, Moldoveanu D, Nguyen TT, Savage P, Rajkumar S, Huang X, Lvova M, Protopopov A, Vuzman D, Hogg D, Park M, Guiot MC, Petrecca K, Mihalcioiu C, Watson IR, Siegel PM, Rose AAN. Dankner M, et al. Clin Cancer Res. 2018 Dec 15;24(24):6483-6494. doi: 10.1158/1078-0432.CCR-17-3384. Epub 2018 Jun 14. Clin Cancer Res. 2018. PMID: 29903896 - Metastatic Melanoma Patient-Derived Xenografts Respond to MDM2 Inhibition as a Single Agent or in Combination with BRAF/MEK Inhibition.
Shattuck-Brandt RL, Chen SC, Murray E, Johnson CA, Crandall H, O'Neal JF, Al-Rohil RN, Nebhan CA, Bharti V, Dahlman KB, Ayers GD, Yan C, Kelley MC, Kauffmann RM, Hooks M, Grau A, Johnson DB, Vilgelm AE, Richmond A. Shattuck-Brandt RL, et al. Clin Cancer Res. 2020 Jul 15;26(14):3803-3818. doi: 10.1158/1078-0432.CCR-19-1895. Epub 2020 Mar 31. Clin Cancer Res. 2020. PMID: 32234759 Free PMC article. - Primary cross-resistance to BRAFV600E-, MEK1/2- and PI3K/mTOR-specific inhibitors in BRAF-mutant melanoma cells counteracted by dual pathway blockade.
Penna I, Molla A, Grazia G, Cleris L, Nicolini G, Perrone F, Picciani B, Del Vecchio M, de Braud F, Mortarini R, Anichini A. Penna I, et al. Oncotarget. 2016 Jan 26;7(4):3947-65. doi: 10.18632/oncotarget.6600. Oncotarget. 2016. PMID: 26678033 Free PMC article. - Clinical Development of BRAF plus MEK Inhibitor Combinations.
Subbiah V, Baik C, Kirkwood JM. Subbiah V, et al. Trends Cancer. 2020 Sep;6(9):797-810. doi: 10.1016/j.trecan.2020.05.009. Epub 2020 Jun 13. Trends Cancer. 2020. PMID: 32540454 Review. - Mitogen-activated protein kinase (MEK) inhibitors to treat melanoma alone or in combination with other kinase inhibitors.
Faghfuri E, Nikfar S, Niaz K, Faramarzi MA, Abdollahi M. Faghfuri E, et al. Expert Opin Drug Metab Toxicol. 2018 Mar;14(3):317-330. doi: 10.1080/17425255.2018.1432593. Epub 2018 Jan 30. Expert Opin Drug Metab Toxicol. 2018. PMID: 29363351 Review.
Cited by
- Targeting Extracellular Matrix Remodeling Restores BRAF Inhibitor Sensitivity in BRAFi-resistant Melanoma.
Marusak C, Thakur V, Li Y, Freitas JT, Zmina PM, Thakur VS, Chang M, Gao M, Tan J, Xiao M, Lu Y, Mills GB, Flaherty K, Frederick DT, Miao B, Sullivan RJ, Moll T, Boland GM, Herlyn M, Zhang G, Bedogni B. Marusak C, et al. Clin Cancer Res. 2020 Nov 15;26(22):6039-6050. doi: 10.1158/1078-0432.CCR-19-2773. Epub 2020 Aug 20. Clin Cancer Res. 2020. PMID: 32820016 Free PMC article. - Inhibition of HGF/MET signaling decreases overall tumor burden and blocks malignant conversion in Tpl2-related skin cancer.
Bonan NF, Kowalski D, Kudlac K, Flaherty K, Gwilliam JC, Falkenberg LG, Maradiaga E, DeCicco-Skinner KL. Bonan NF, et al. Oncogenesis. 2019 Jan 10;8(1):1. doi: 10.1038/s41389-018-0109-8. Oncogenesis. 2019. PMID: 30631034 Free PMC article. - Patient-Derived Xenograft Models of Non-Small Cell Lung Cancer and Their Potential Utility in Personalized Medicine.
Morgan KM, Riedlinger GM, Rosenfeld J, Ganesan S, Pine SR. Morgan KM, et al. Front Oncol. 2017 Jan 19;7:2. doi: 10.3389/fonc.2017.00002. eCollection 2017. Front Oncol. 2017. PMID: 28154808 Free PMC article. - Response and Resistance to Paradox-Breaking BRAF Inhibitor in Melanomas In Vivo and Ex Vivo.
Hartsough EJ, Kugel CH 3rd, Vido MJ, Berger AC, Purwin TJ, Goldberg A, Davies MA, Schiewer MJ, Knudsen KE, Bollag G, Aplin AE. Hartsough EJ, et al. Mol Cancer Ther. 2018 Jan;17(1):84-95. doi: 10.1158/1535-7163.MCT-17-0705. Epub 2017 Nov 13. Mol Cancer Ther. 2018. PMID: 29133617 Free PMC article. - Patient-derived xenografts: a relevant preclinical model for drug development.
Pompili L, Porru M, Caruso C, Biroccio A, Leonetti C. Pompili L, et al. J Exp Clin Cancer Res. 2016 Dec 5;35(1):189. doi: 10.1186/s13046-016-0462-4. J Exp Clin Cancer Res. 2016. PMID: 27919280 Free PMC article. Review.
References
- Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372:30–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- P01 CA025874/CA/NCI NIH HHS/United States
- P01 CA114046/CA/NCI NIH HHS/United States
- R01 CA174746/CA/NCI NIH HHS/United States
- P50 CA174523/CA/NCI NIH HHS/United States
- T32 CA078207/CA/NCI NIH HHS/United States
- R01 CA174746-01/CA/NCI NIH HHS/United States
- R01 CA047159/CA/NCI NIH HHS/United States
- P30CA010815/CA/NCI NIH HHS/United States
- P30 CA010815/CA/NCI NIH HHS/United States
- P30CA016672/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous