Identification of key pathways and metabolic fingerprints of longevity in C. elegans - PubMed (original) (raw)
Identification of key pathways and metabolic fingerprints of longevity in C. elegans
Arwen W Gao et al. Exp Gerontol. 2018 Nov.
Abstract
Impaired insulin/IGF-1 signaling (IIS) and caloric restriction (CR) prolong lifespan in the nematode C. elegans. However, a cross comparison of these longevity pathways using a multi-omics integration approach is lacking. In this study, we aimed to identify key pathways and metabolite fingerprints of longevity that are shared between IIS and CR worm models using multi-omics integration. We generated transcriptomics and metabolomics data from long-lived worm strains, i.e. daf-2 (impaired IIS) and eat-2 (CR model) and compared them with the wild-type strain N2. Transcriptional profiling identified shared longevity signatures, such as an upregulation of lipid storage and defense responses, and downregulation of macromolecule synthesis and developmental processes. Metabolomics profiling identified an increase in the levels of glycerol‑3P, adenine, xanthine, and AMP, and a decrease in the levels of the amino acid pool, as well as the C18:0, C17:1, C19:1, C20:0 and C22:0 fatty acids. After we integrated transcriptomics and metabolomics data based on the annotations in KEGG, our results highlighted increased amino acid metabolism and an upregulation of purine metabolism as a commonality between the two long-lived mutants. Overall, our findings point towards the existence of shared metabolic pathways that are likely important for lifespan extension and provide novel insights into potential regulators and metabolic fingerprints for longevity.
Keywords: C. elegans; Caloric restriction; Insulin/IGF-1 signaling; Longevity; Metabolomics; Transcriptomics.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
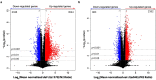
Fig. 1
Transcriptional alterations in long-lived daf-2 and eat-2 mutants compared to N2 worms. (a) Volcano plot of log2-transformed fold change of gene expression in daf-2(e1370) over N2 worms. (b) Volcano plot of log2-transformed fold change of gene expression in eat-2(ad465) compared to those in N2 worms. _p_-Value was calculated using Student's _t_-test. The vertical dashed lines represent the log2 fold change = −1 or 1, horizontal dashed lines denote _p_-value cutoffs. The number of genes that are significantly up- (red) or down- (blue) regulated (_p_-value < 0.05) in each panel is also mentioned. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
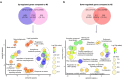
Fig. 2
Venn diagram and GO term enrichment analysis comparing daf-2 and eat-2 mutants with N2. (a) Venn diagram of significantly upregulated genes in both long-lived mutants identified 1722 shared transcripts. GO term enrichment (biological process) of these genes using David and ReviGO highlighted several metabolic pathways such as lipid metabolism and phosphorus metabolic process. Venn diagram percentages in brackets indicate the number of genes that account for the % of the total up-/downregulated genes. (b) Venn diagram of significantly downregulated genes in both long-lived mutants identified 617 transcripts. GO term enrichment (biological process) of these genes using David and ReviGO shows an attenuation of process that are involved in biosynthesis of macromolecules such as aromatic compounds, nucleobase-containing compounds and nitrogen compounds. The size of the plot is indicative of the frequency of the GO term in the underlying Gene Ontology Annotation database; the plots are color-coded according to significance (log10-transformed); level of significance increases from red to blue. GO terms belonging to the same cluster were grouped and circled in dark grey dashed line. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
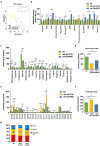
Fig. 3
Metabolite signatures of the long-lived mutants daf-2 and eat-2. (a) Principle Component Analysis (PCA) analysis score plot showing group separation based on polar metabolite profiles in both long-lived mutants compared to N2 worms, ellipses represent the 95% confidence interval. (b) Polar metabolite profiles of daf-2 and eat-2 mutants compared to N2 worms (metabolites that are significantly different (FDR < 0.05) are selected and the _p_-values from the _t_-test is indicated as *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). Several shared metabolic signatures were detected, including increased levels of AMP, decreased levels of glycerol‑3P, adenine, and xanthine. Unique metabolite signatures for each long-lived mutant were also observed, for the daf-2 mutant: increased levels of phosphoenolpyruvate, and decreased levels of citrate/isocitrate, gluconate, ADP, succinate and malate; for the eat-2 mutant: increased levels of succinate, UMP, uracil, GMP, and ADP. (c) Most amino acid species in both long-lived mutants were lowered compared to N2 worms. (d) Total amino acids in both daf-2 and eat-2 mutants were significantly lower compared to that in N2 worms. (e) A number of abundant fatty acids were increased in daf-2 mutants compared to N2 worms, whereas most fatty acids in eat-2 mutants were decreased compared to N2. (f) Total fatty acids in daf-2 mutants were significantly elevated while those in eat-2 mutants were significantly decreased. (g) Mono-unsaturated fatty acids (MUFA) were more abundant in daf-2 mutants and PUFAs were more abundant in eat-2 mutants. The bar graphs depict mean ± SD. Significance in panel c–f was calculated using one-way ANOVA; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
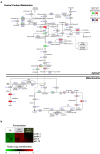
Fig. 4
Central carbon metabolism pathways in daf-2 and eat-2 mutants. (a) Data from the gene expression profile (rectangles) and metabolite profile (circles) were color-coded and integrated in three major pathways of central carbon metabolism, including glycolysis/gluconeogenesis, the pentose phosphate pathway and TCA cycle. The majority of genes involved in glycolysis/gluconeogenesis remained unaltered in both long-lived mutants. Glycolysis/gluconeogenesis intermediates remained low in both long-lived mutants, except the final product of this pathway, i.e. pyruvate, which accumulated in daf-2 mutants. Genes coding for PPP enzymes were mostly unchanged and PPP intermediates were lowered in both long-lived mutants. Alcohol fermenting genes also showed increased expression in both long-lived mutants. The majority of genes involved in the TCA cycle were unchanged and TCA intermediates were significantly lowered in daf-2 mutants. TCA intermediates were unchanged in eat-2 mutants except for elevated _cis_‑aconitate and succinate levels. White circles are metabolites that were not measured. (b) Gene expression profiles of genes that are involved in fermentative metabolism. Each column represents one biological replicate (four biological replicates for each worm strain), and the fold-change is represented as log2-transformed.
Fig. 5
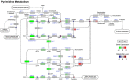
Pyrimidine metabolism pathways in daf-2 and eat-2 mutants. Genes involved in the pyrimidine metabolism in both long-lived mutants were significantly down-regulated compared to those in N2 worms. Only a few genes involved in pyrimidine degradation were upregulated. In line with the expression, most pyrimidine intermediates were low abundant in both long-lived mutants. White circles are metabolites that were not measured.
Fig. 6
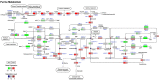
Purine metabolism pathways of daf-2 and eat-2 mutants. The majority of genes involved in the three major pathways (de novo synthesis, salvage, and degradation) of purine metabolism were significantly upregulated in both daf-2 and eat-2. Three metabolites showed similar changes in both mutants, i.e. AMP, adenine, and xanthine. Most purine metabolism intermediates were decreased in daf-2 mutants, except guanine; in eat-2 mutants the differences in purine intermediate abundance is more diverse. White circles are metabolites that were not measured.
Fig. 7
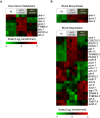
Heat maps showing transcriptional changes in specific gene sets. Gene expression profiles of gene sets that are involved in one carbon metabolism (a), and branched-chain amino acid (BCAA) metabolism (b). Each column represents one biological replicate (four biological replicates for each worm strain), and the fold-change is represented as log2-transformed.
Similar articles
- Global Cysteine-Reactivity Profiling during Impaired Insulin/IGF-1 Signaling in C. elegans Identifies Uncharacterized Mediators of Longevity.
Martell J, Seo Y, Bak DW, Kingsley SF, Tissenbaum HA, Weerapana E. Martell J, et al. Cell Chem Biol. 2016 Aug 18;23(8):955-66. doi: 10.1016/j.chembiol.2016.06.015. Epub 2016 Aug 4. Cell Chem Biol. 2016. PMID: 27499530 Free PMC article. - Insulin/IGF-1-mediated longevity is marked by reduced protein metabolism.
Stout GJ, Stigter EC, Essers PB, Mulder KW, Kolkman A, Snijders DS, van den Broek NJ, Betist MC, Korswagen HC, Macinnes AW, Brenkman AB. Stout GJ, et al. Mol Syst Biol. 2013 Jul 2;9:679. doi: 10.1038/msb.2013.35. Mol Syst Biol. 2013. PMID: 23820781 Free PMC article. - ATGL-1 mediates the effect of dietary restriction and the insulin/IGF-1 signaling pathway on longevity in C. elegans.
Zaarur N, Desevin K, Mackenzie J, Lord A, Grishok A, Kandror KV. Zaarur N, et al. Mol Metab. 2019 Sep;27:75-82. doi: 10.1016/j.molmet.2019.07.001. Epub 2019 Jul 5. Mol Metab. 2019. PMID: 31311719 Free PMC article. - The longevity effect of dietary restriction in Caenorhabditis elegans.
Houthoofd K, Vanfleteren JR. Houthoofd K, et al. Exp Gerontol. 2006 Oct;41(10):1026-31. doi: 10.1016/j.exger.2006.05.007. Epub 2006 Jun 19. Exp Gerontol. 2006. PMID: 16782293 Review. - The role of insulin/IGF-1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster.
Altintas O, Park S, Lee SJ. Altintas O, et al. BMB Rep. 2016 Feb;49(2):81-92. doi: 10.5483/bmbrep.2016.49.2.261. BMB Rep. 2016. PMID: 26698870 Free PMC article. Review.
Cited by
- Targeting metabolic pathways for extension of lifespan and healthspan across multiple species.
Parkhitko AA, Filine E, Mohr SE, Moskalev A, Perrimon N. Parkhitko AA, et al. Ageing Res Rev. 2020 Dec;64:101188. doi: 10.1016/j.arr.2020.101188. Epub 2020 Oct 5. Ageing Res Rev. 2020. PMID: 33031925 Free PMC article. Review. - The Native Microbiome Member Chryseobacterium sp. CHNTR56 MYb120 Induces Trehalose Production via a Shift in Central Carbon Metabolism during Early Life in C. elegans.
Shiri TJ, Viau C, Gu X, Xu L, Lu Y, Xia J. Shiri TJ, et al. Metabolites. 2023 Aug 18;13(8):953. doi: 10.3390/metabo13080953. Metabolites. 2023. PMID: 37623896 Free PMC article. - Genetic analysis of Caenorhabditis elegans pry-1/Axin suppressors identifies genes involved in reproductive structure development, stress responses, and aging.
Mallick A, Jhaveri N, Jeon J, Chang Y, Shah K, Hosein H, Gupta BP. Mallick A, et al. G3 (Bethesda). 2022 Feb 4;12(2):jkab430. doi: 10.1093/g3journal/jkab430. G3 (Bethesda). 2022. PMID: 35100345 Free PMC article. - Age-related changes of skeletal muscle metabolic response to contraction are also sex-dependent.
Campbell MD, Djukovic D, Raftery D, Marcinek DJ. Campbell MD, et al. J Physiol. 2025 Jan;603(1):69-86. doi: 10.1113/JP285124. Epub 2023 Sep 23. J Physiol. 2025. PMID: 37742081 Free PMC article. - Differential microRNAs expression profiles in liver from three different lifestyle modification mice models.
Gong H, Zhang M, Han Y, Zhang Y, Pang J, Zhao Y, Chen B, Wu W, Qi R, Zhang T. Gong H, et al. BMC Genomics. 2021 Mar 19;22(1):196. doi: 10.1186/s12864-021-07507-3. BMC Genomics. 2021. PMID: 33740891 Free PMC article.
References
- Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous